New tools for popgen simulation and analysis:
What’s possible?
Peter Ralph
University of Oregon
Institute of Ecology and Evolution
Evolution 2021
slides: github:petrelharp/evolution_2021
Overview of simulators
In this talk:
Forwards or backwards?
Forwards or backwards?
Do your digital organisms:
- have at most one site under selection?
- live in a collection of randomly-mating populations?
- not need some specific life cycle?
If so, then coalescent simulation is the way to go!
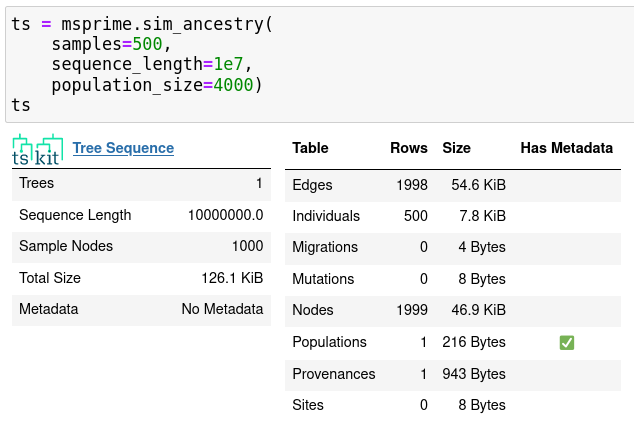
msprime


msprime v1.0

New features:
- \(k\)-ploid individuals, finite sites
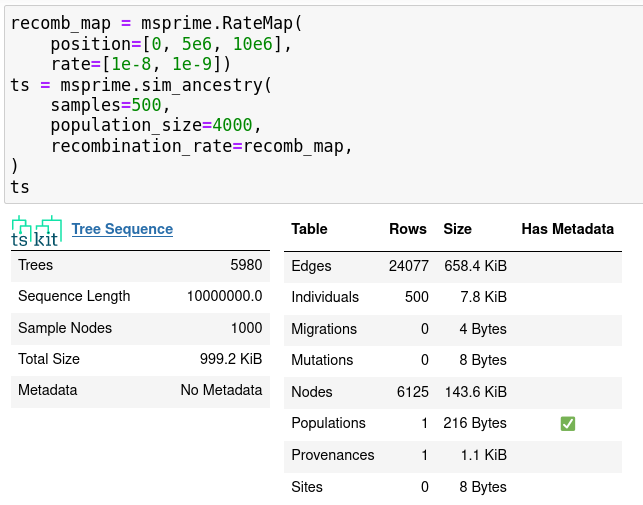
- recombination rate maps
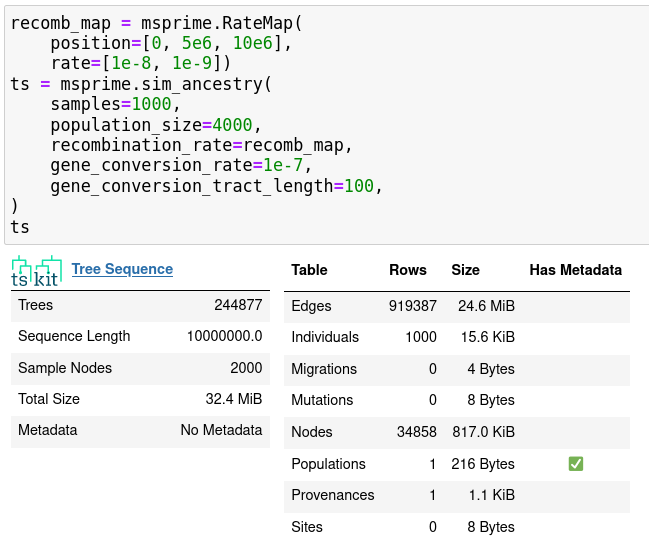
- gene conversion
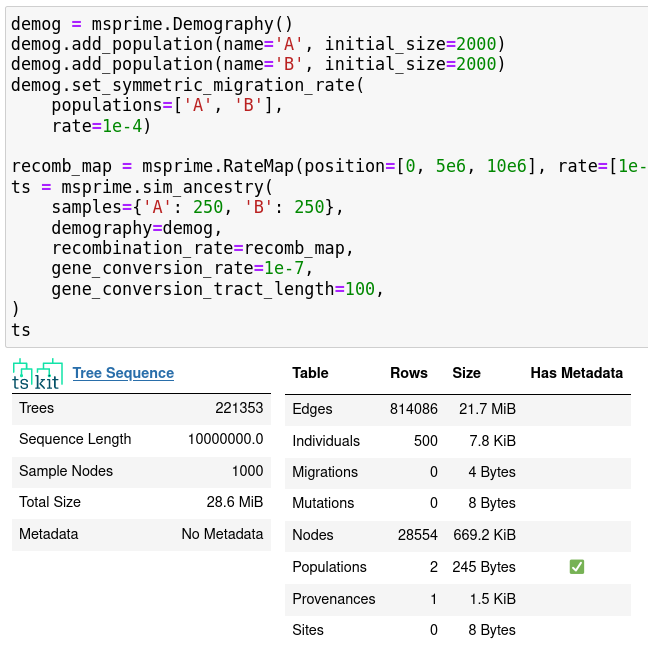
- nicer demographic model specification
- mutation rate maps
- quick analysis
New features:
- \(k\)-ploid individuals, finite sites
- recombination rate maps
- gene conversion
- nicer demographic model specification
- mutation rate maps
- quick analysis
New features:
- \(k\)-ploid individuals, finite sites
- recombination rate maps
- gene conversion
- nicer demographic model specification
- mutation rate maps
- quick analysis
New features:
- \(k\)-ploid individuals, finite sites
- recombination rate maps
- gene conversion
- nicer demographic model specification
- mutation rate maps
- quick analysis
New features:
- \(k\)-ploid individuals, finite sites
- recombination rate maps
- gene conversion
- nicer demographic model specification
- mutation rate maps
- quick analysis
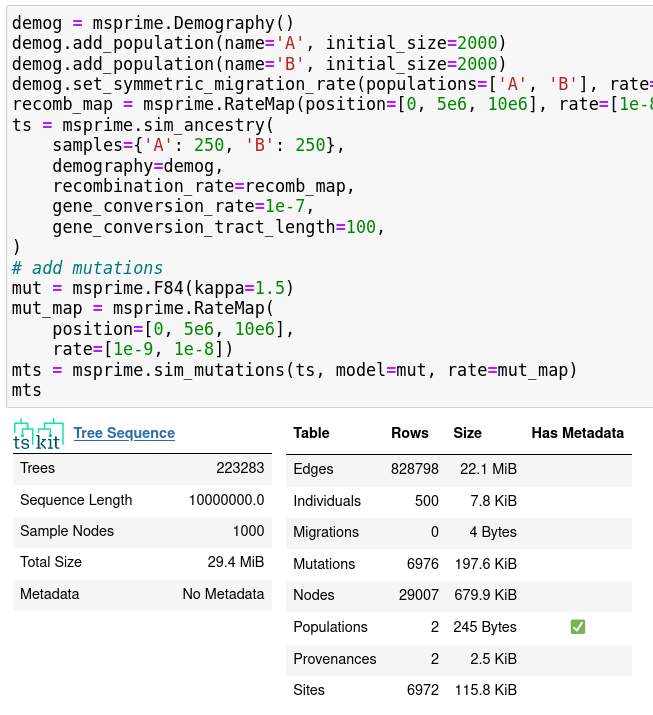
New features:
- \(k\)-ploid individuals, finite sites
- recombination rate maps
- gene conversion
- nicer demographic model specification
- mutation rate maps
- quick analysis
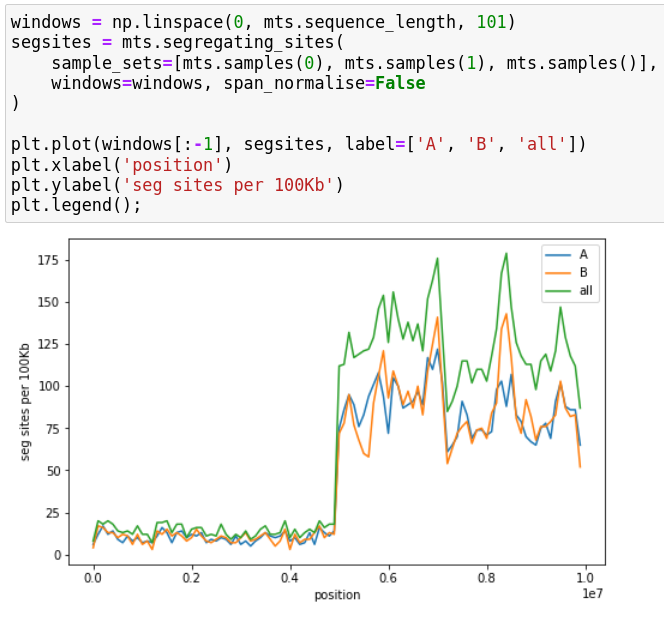
Ancestry models
- “the” coalescent
- discrete-time Wright-Fisher
- multiple mergers
- selective sweeps
sweep_model = msprime.SweepGenicSelection(
position=2.5e4, s=0.01,
start_frequency=0.5e-4, end_frequency=0.99, dt=1e-6)
sts = msprime.sim_ancestry(9,
model=[sweep_model, msprime.StandardCoalescent()],
population_size=1e4, recombination_rate=1e-8, sequence_length=5e4)Mutation models
- infinite sites/alleles
- nucleotides
- amino acids
- arbitrary Markovian models
dem = msprime.Demography.from_species_tree(
"((A:900,B:900)ab:100,C:1000)abc;",
initial_size=1e3)
samples = {"A": 2, "B": 1, "C": 1}
ts = msprime.sim_ancestry(
8, demography=dem, sequence_length=5e4,
recombination_rate=1e-8
)
mts = msprime.sim_mutations(ts, rate=1e-7)
mts.draw_svg()SLiMv3
An eco-evolutionary simulator
- everything msprime can
- ecological dynamics with “non-Wright-Fisher” models
- populations in continuous, heterogeneous geography
- sex chromosomes, haplodiploidy
- complex traits
- context-dependent mutations
- v4: interacting species

Ben Haller
Getting started:
- read the introduction of the SLiM manual
- find a recipe that’s close to what you want
- open up the GUI and try it
- print stuff in the console
- add in other bits
- take a workshop!
tree sequences



Development philosophy
- open, welcoming, supportive
- well-documented
- reliable, reproducible
- backwards compatible
The tree sequence
video credit: Yan Wong
Benefits
Post-hoc mutations
Recapitation
Runtime
- \(N_e\) = population size
- \(L\) = genome length
- \(T\) = # of generations
- sample size doesn’t matter
- “chromosome” = \(10^8\) bp
msprime: quadratic in \(N_e L\)- chromosomes, \(N_e = 1,000\): seconds
- megabases, \(N_e = 100,000\): seconds
- chromosomes, \(N_e = 100,000\): hours
- megabases, \(N_e = 10,000,000\): hours
SLiM: linear in \(N_e T\)- \(N_e = 1,000\): seconds/thousand gens
- \(N_e = 100,000\): minutes/thousand gens
- selection: 3x slower
- space: 10x slower with neighborhood size 20
How long do I run it for?
- Until equilibrium. (4N? 20N?)
- If that’s too long, for a “while”, and recapitate.
- Your results shouldn’t depend too much on how you do it.
Big picture: how accurate do you think your demographic model reflects 2N generations ago, really?
Considerations
- \(N\) = population size
- \(L\) = genome length
- sample size (doesn’t matter much)
- number of generations (SLiM only)
- selection
- geography
- adding neutral mutations (nearly instant)
msprime: 1000 samples
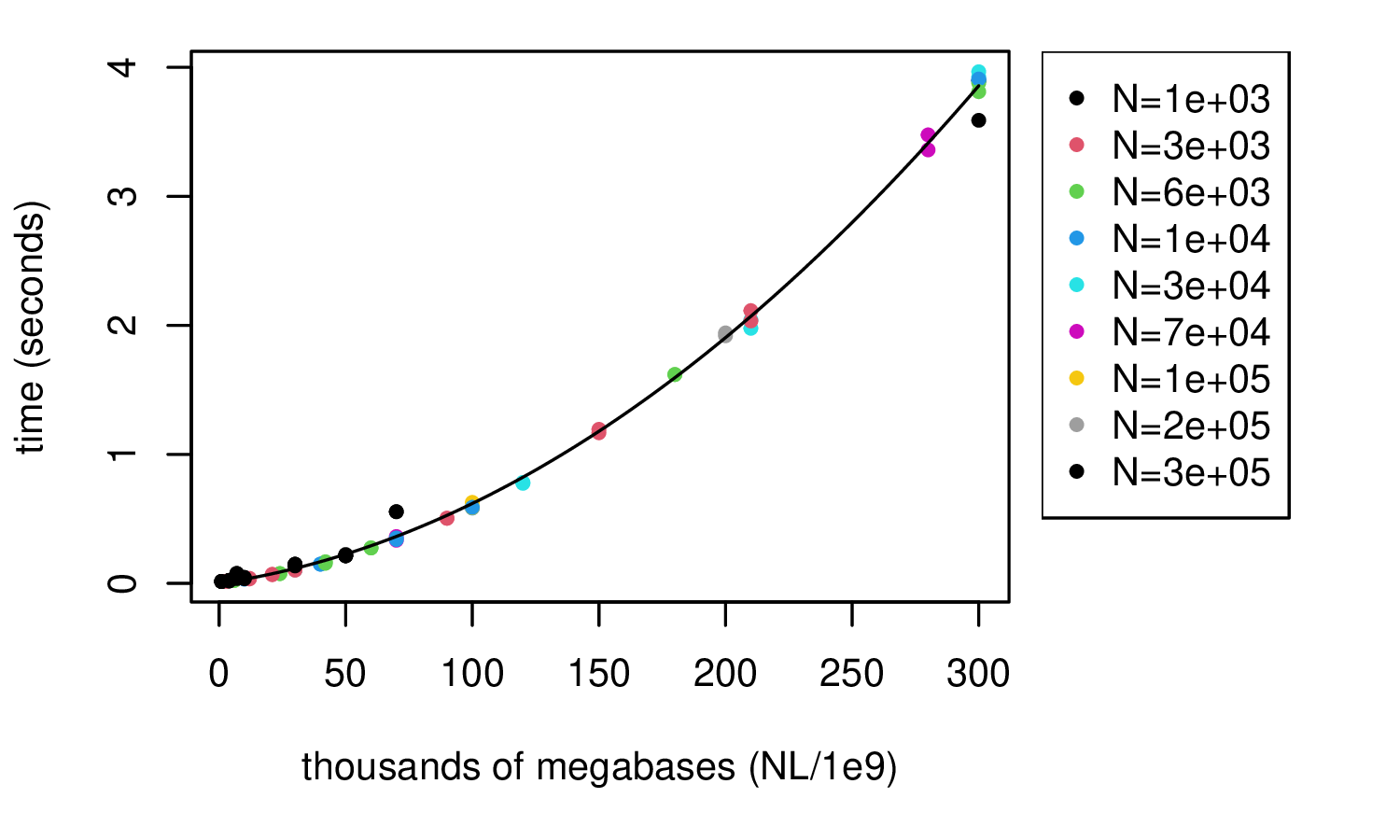
takeaway: hundreds of thousands of megabases takes seconds
msprime: 1000 samples
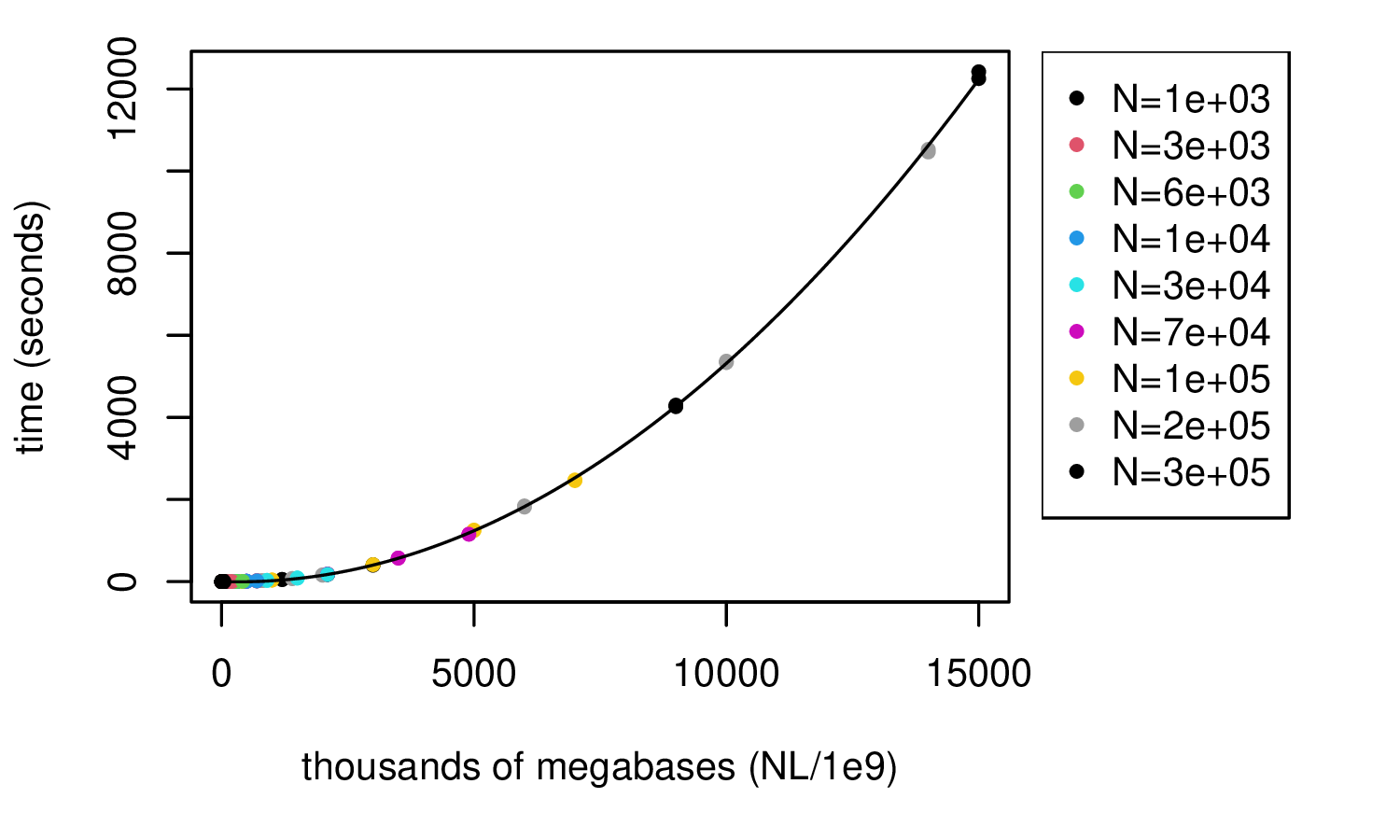
takeaway: hundreds of thousands of megabases takes seconds
basic demography: SLiM
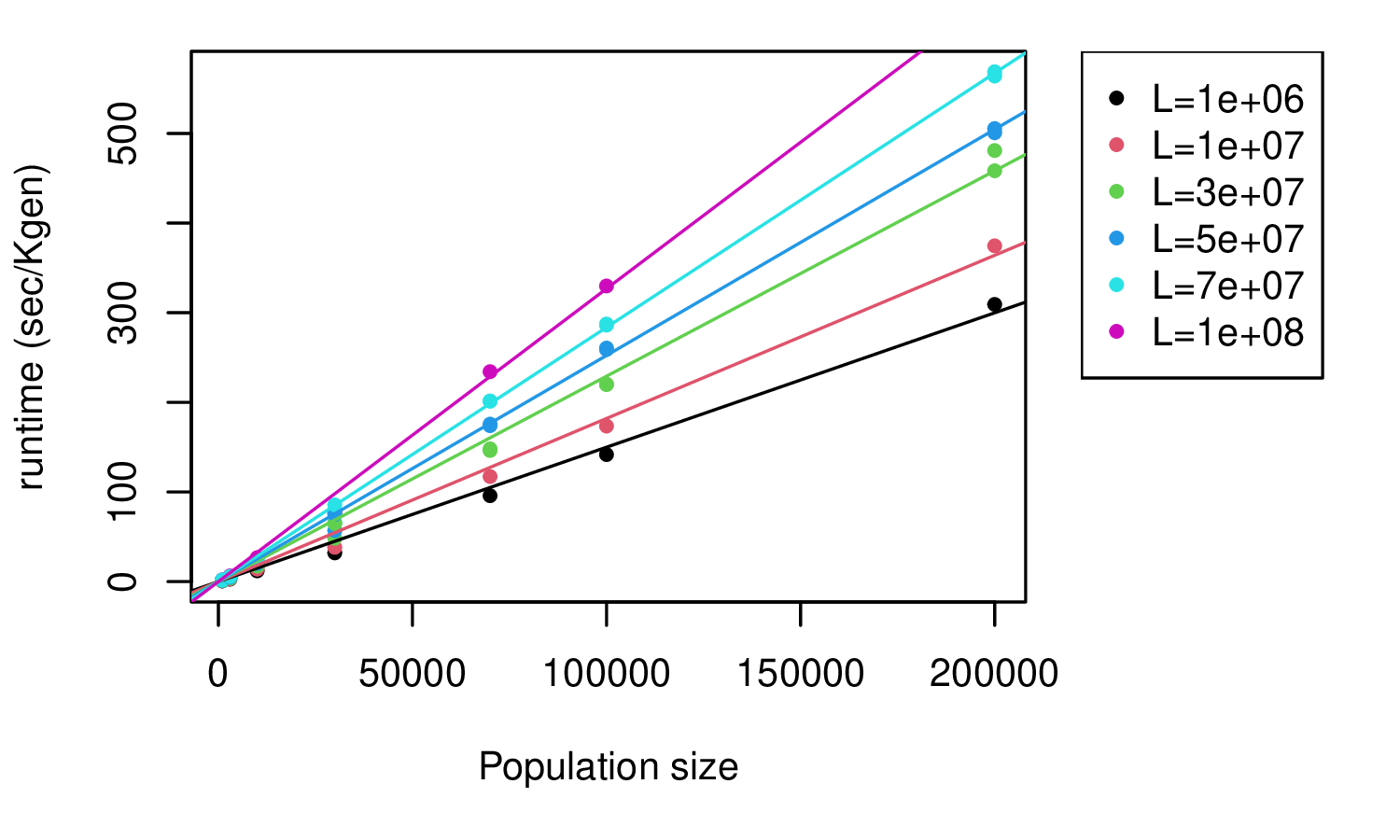
takeaway: linear in population size
Basic demography: SLiM
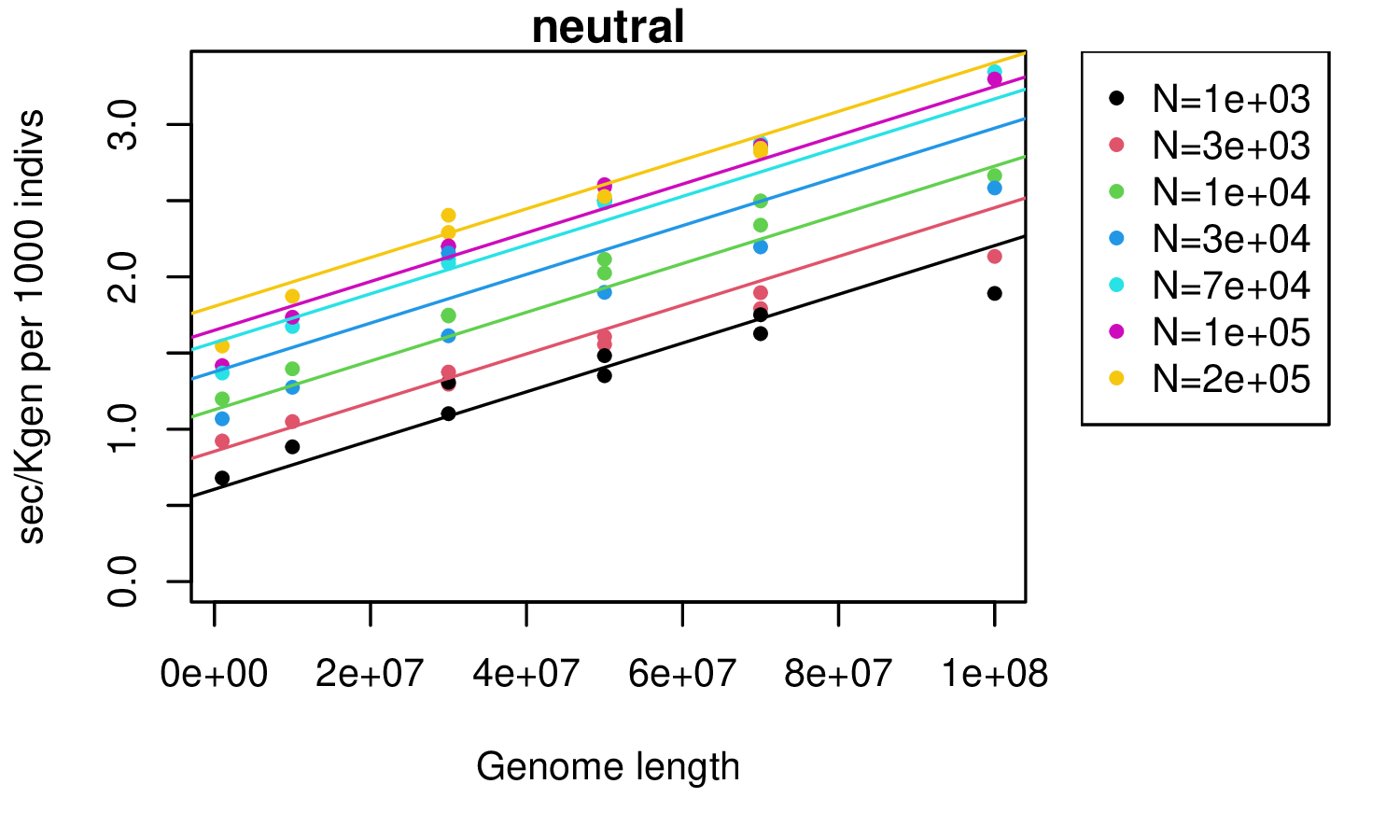
takeaway: seconds per thousand individuals per thousand generations
Selection: SLiM, total rate \(10^{-10}\)
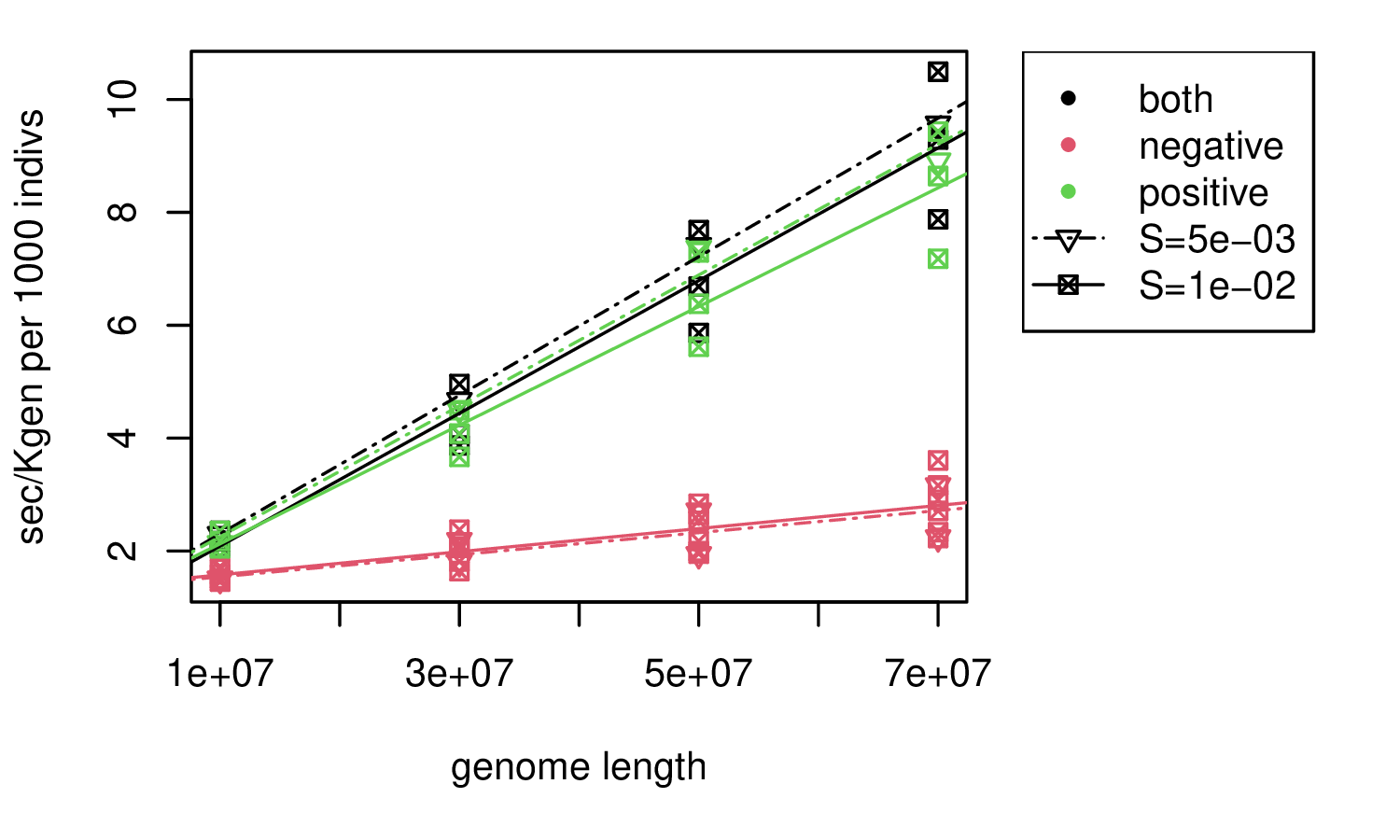
takeaway: similar, but slower by a factor of 3 for lots of positive mutations
Spatial simulations: SLiM
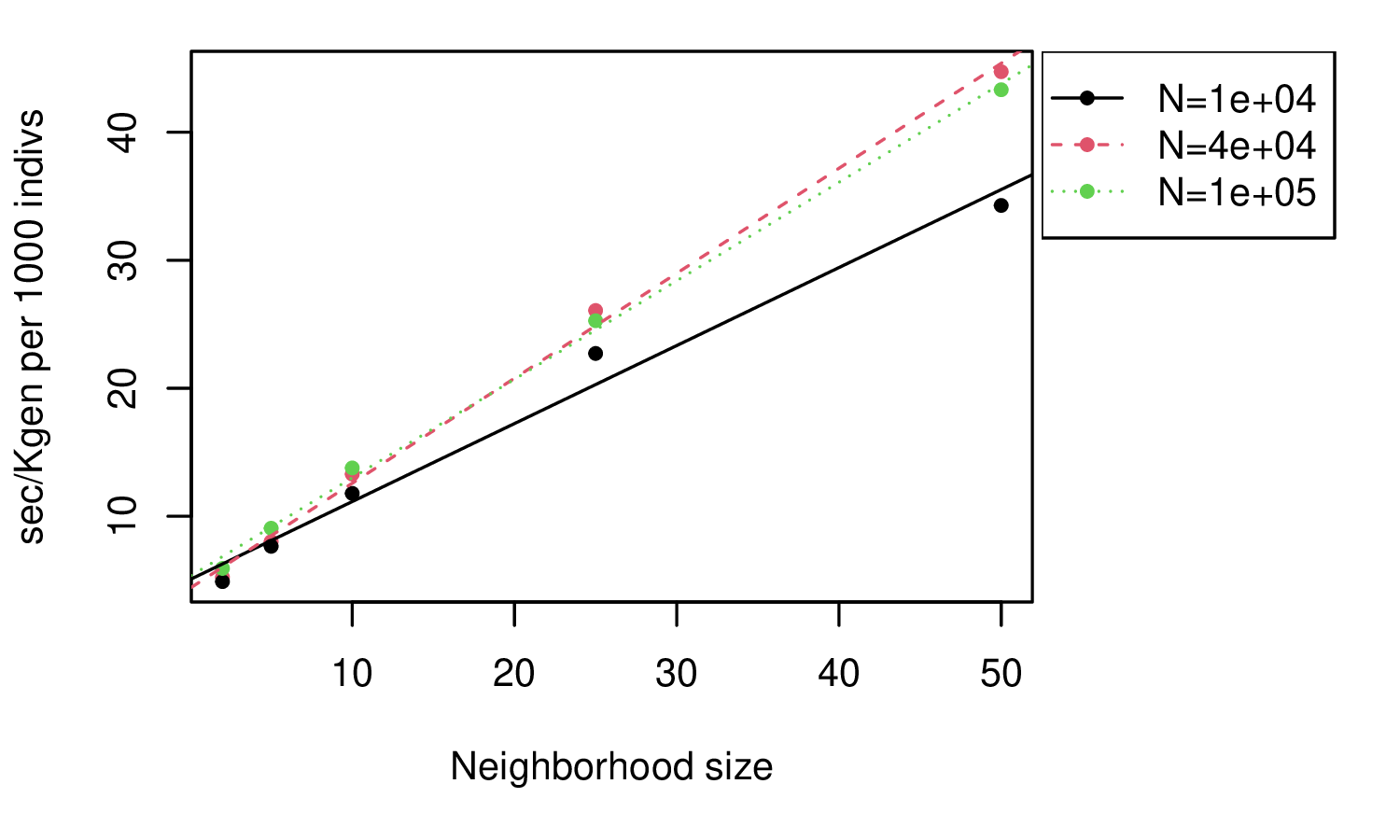
takeaway: 3x slower than genomes! Scales with neighborhood size (\(\sigma^2\)).
Thanks!



- Jerome Kelleher
- Ben Haller
- Ben Jeffery
- Yan Wong
- Murillo Rodrigues
- Andy Kern
- Philipp Messer
How to get help
SLiM: the mailing list
msprime/tskit: “discussions” on github
Get involved! Suggest features, write documentation, write code…