A practical introduction to the tree sequence:
Peter Ralph, University of Oregon
June 2023
Genomes and genealogies
Genomes
- are very big (\(10^7\)–\(10^{12}\) nucleotides)
- reflect past history and process
A data structure is…
succinct if it only stores each bit of information once.
descriptive if it reflects the underlying process.
So: let’s think about the process that generated the data!
Meiosis & Recombination
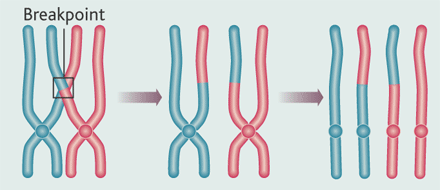
You have two copies of each autosome, one from each parent.
When you make a gamete, the copies recombine,
and copying errors lead to mutations.
Each two copies of the genome were inherited, noisily, from the two parents,
and from the four grandparents,
and the eight great-grandparents
and the sixteen great-great-grandparents
… but, how much from each of them?
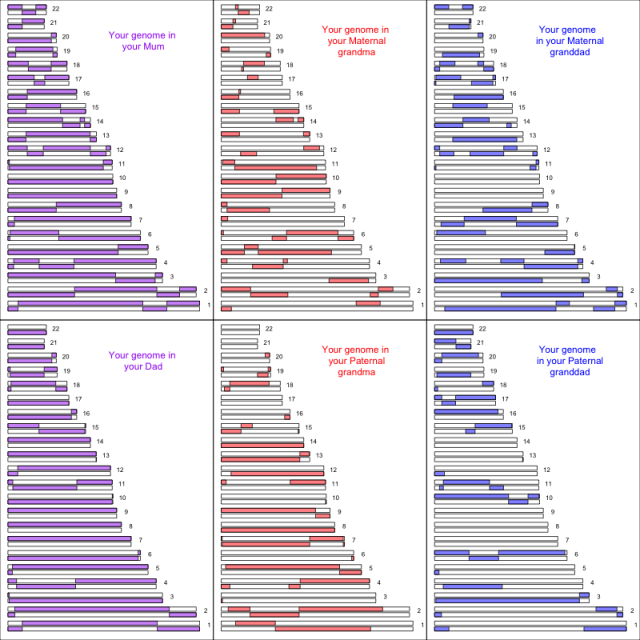
from gcbias.org
Looking backwards
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
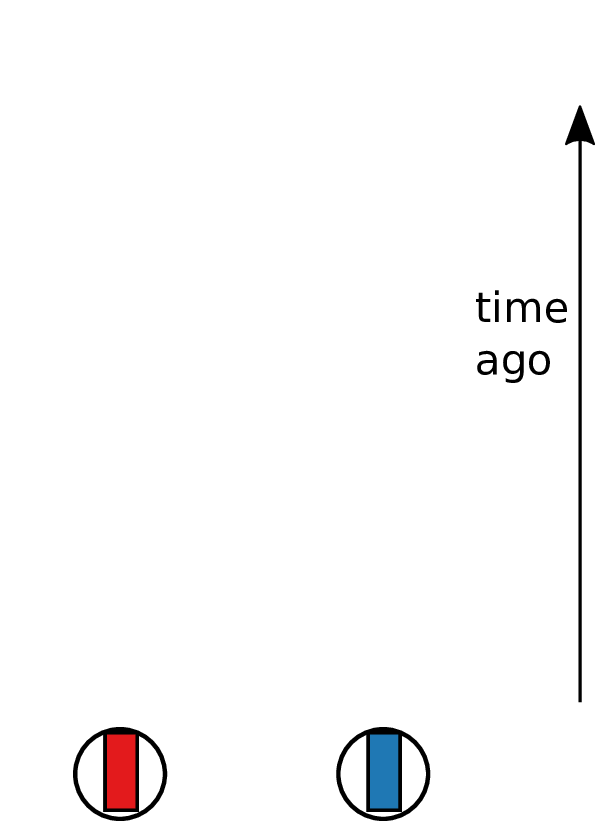
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
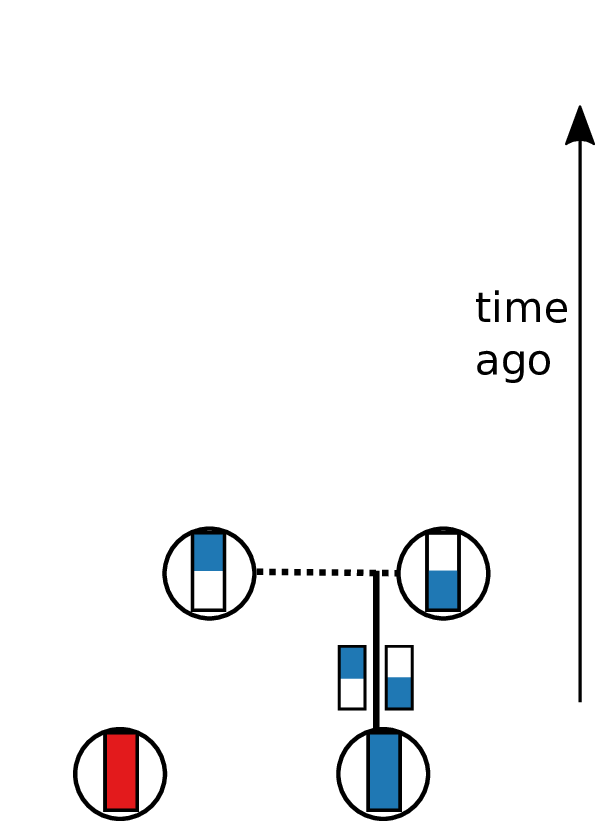
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
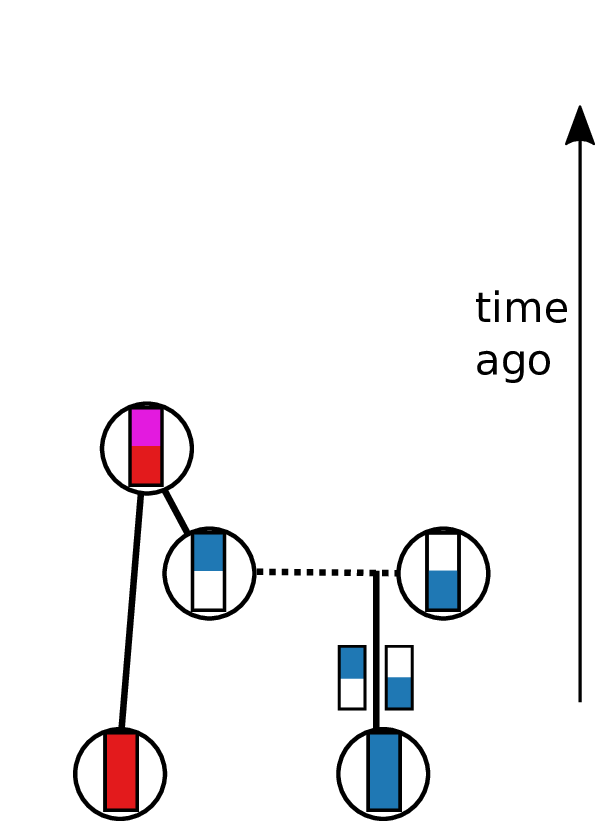
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
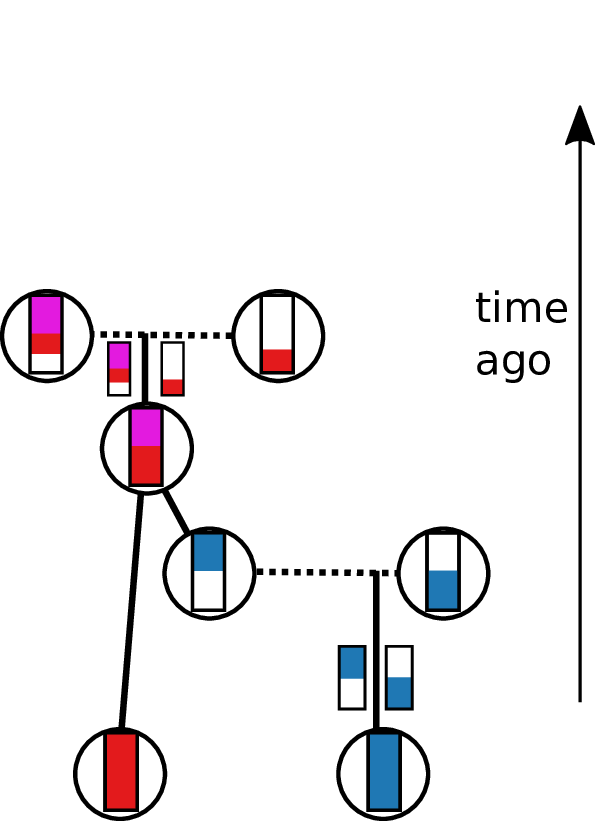
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
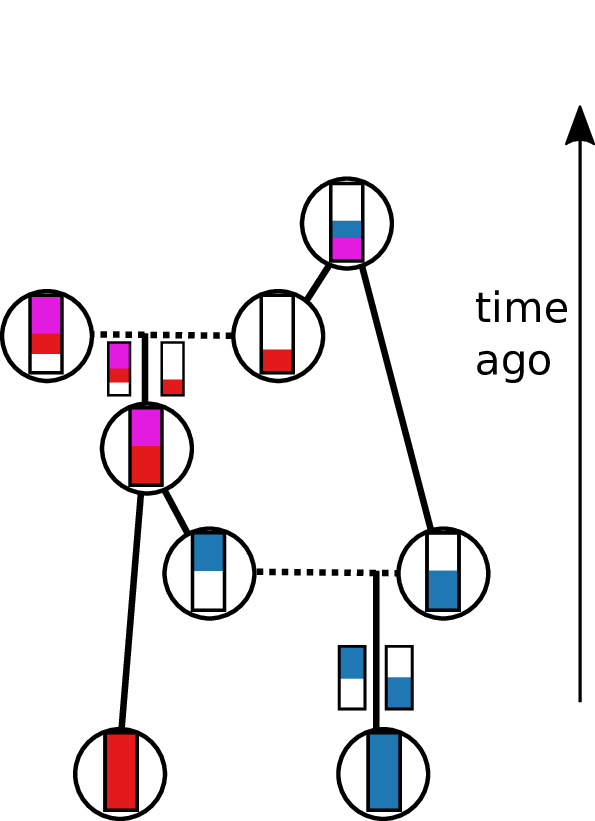
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
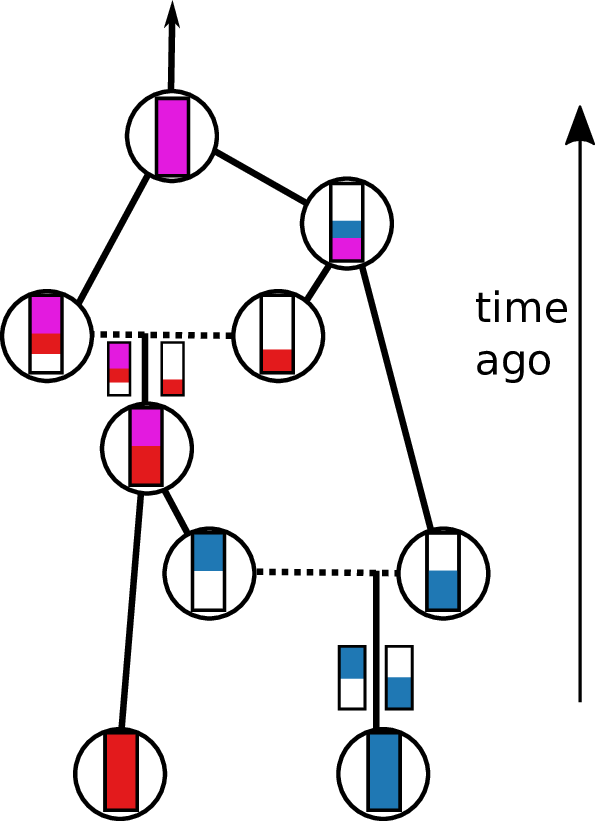
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.
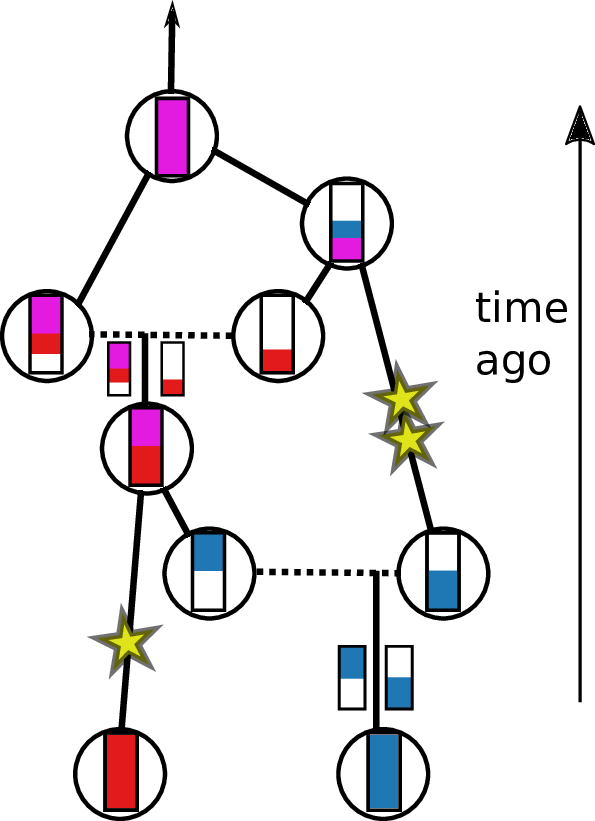
Tracing back the ancestry of some chromosomes:
- blocks labeled by who inherits from them
- blocks can split
- or coalesce,
- and mutations lead to differences.
Result: a labeled genealogy containing all the genealogical trees.

The tree sequence
History is a sequence of trees
For a set of sampled chromosomes, at each position along the genome there is a genealogical tree that says how they are related.
The succinct tree sequence
is a way to succinctly describe this, er, sequence of trees
and the resulting genome sequences.

by Jerome Kelleher, in Kelleher, Etheridge, and McVean
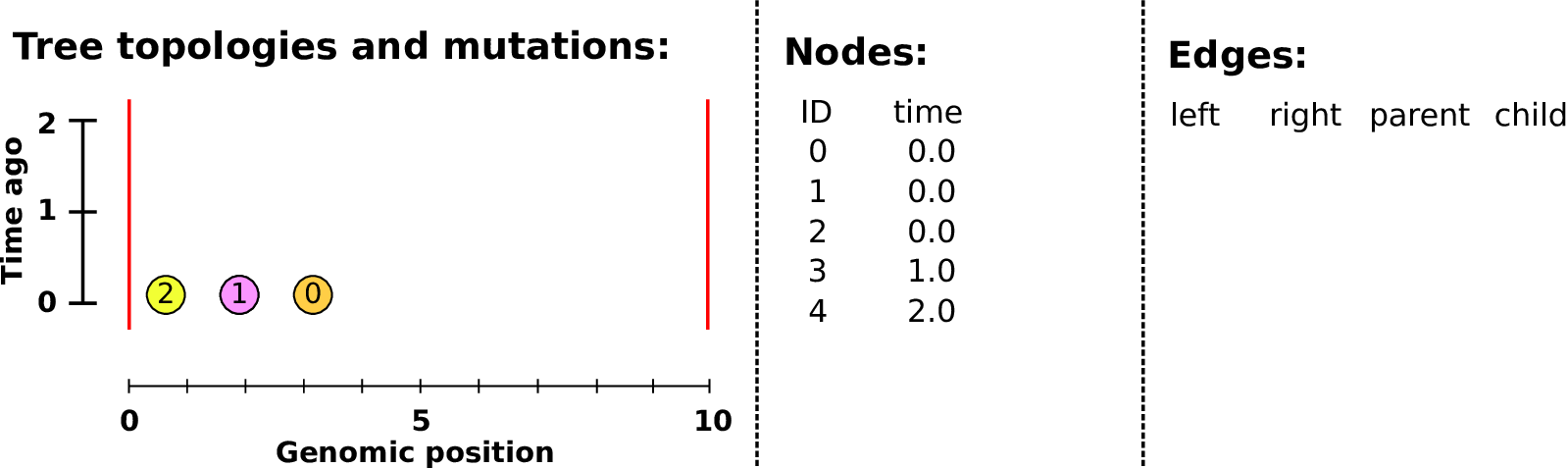
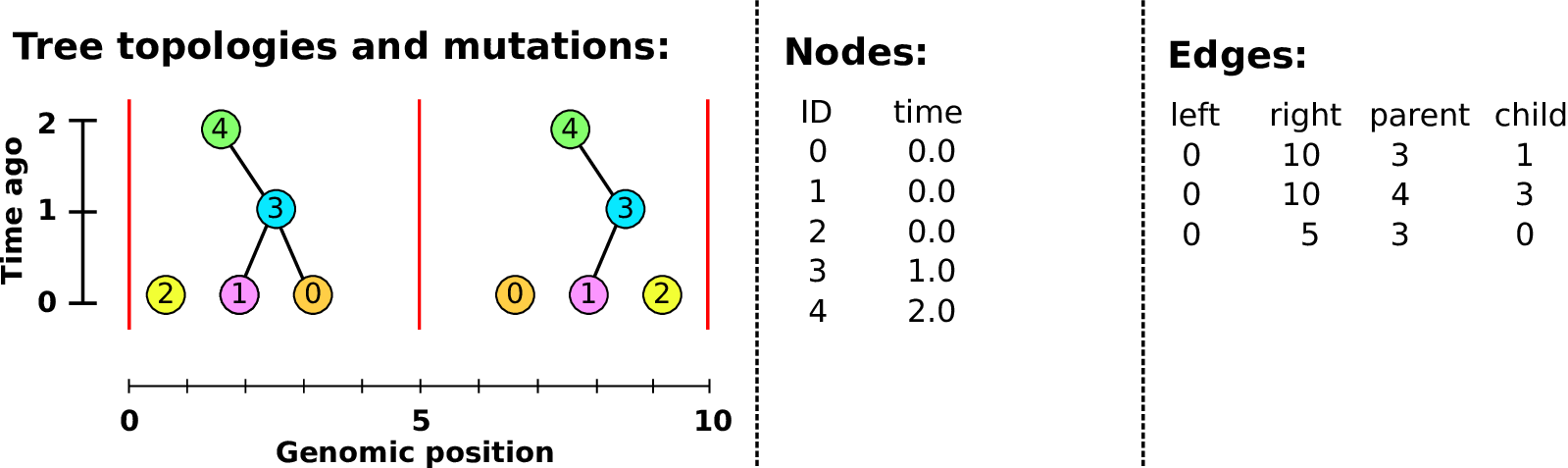
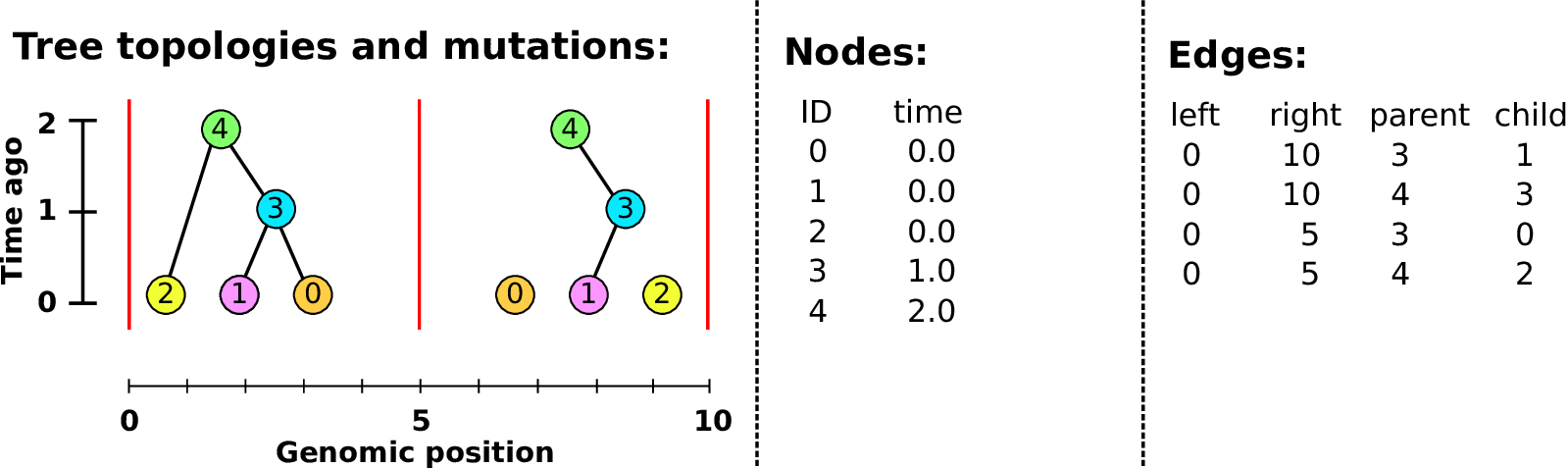
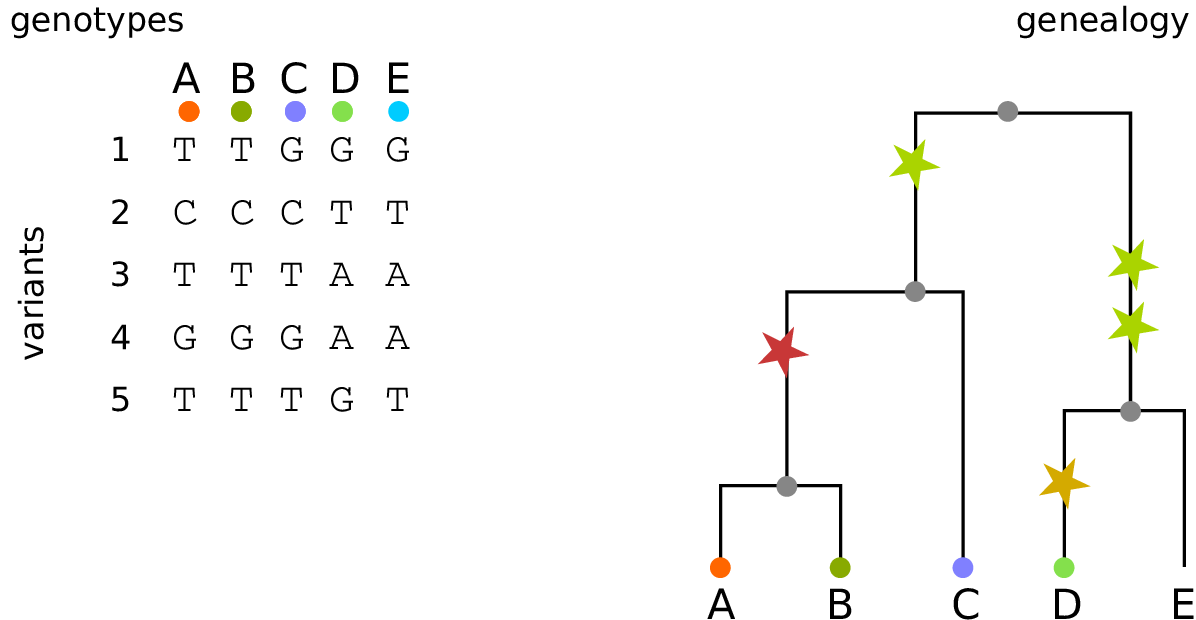
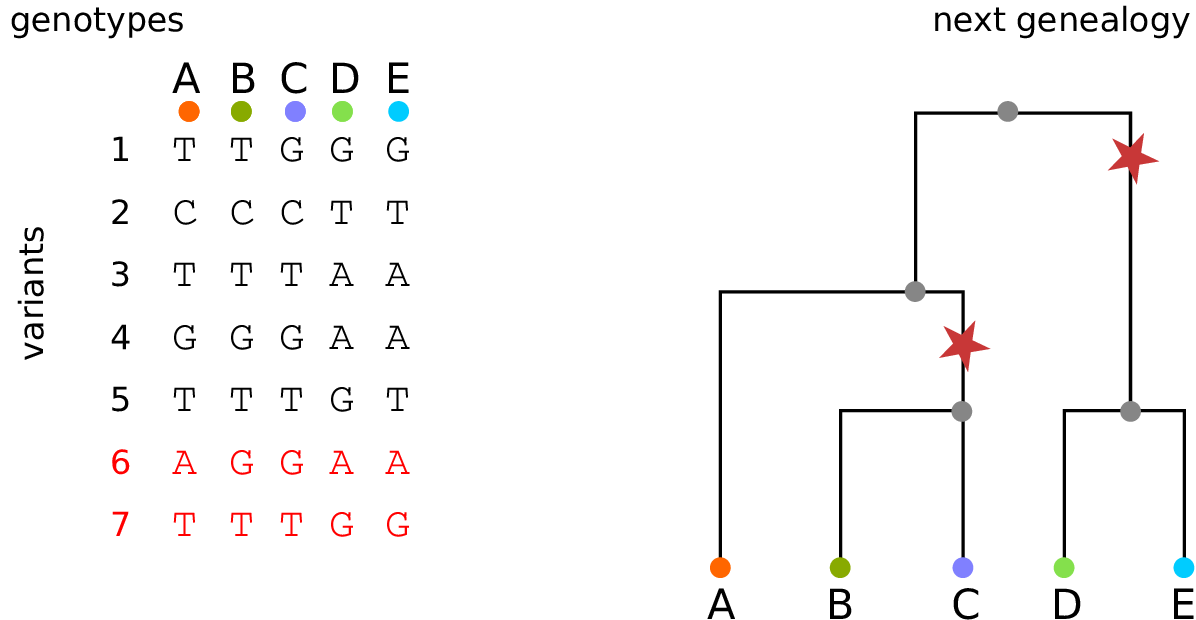
Example: three samples; two trees; two variant sites
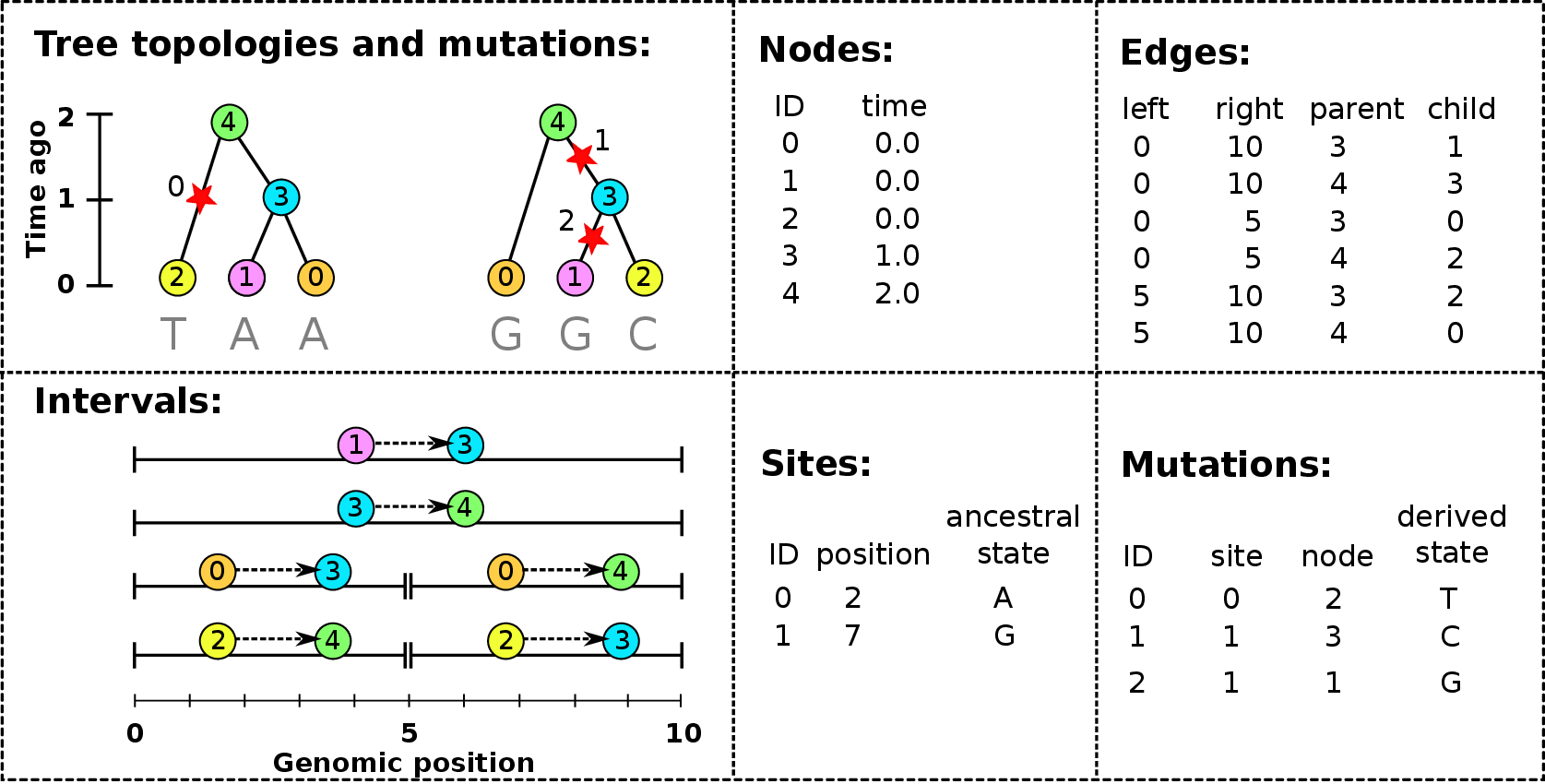
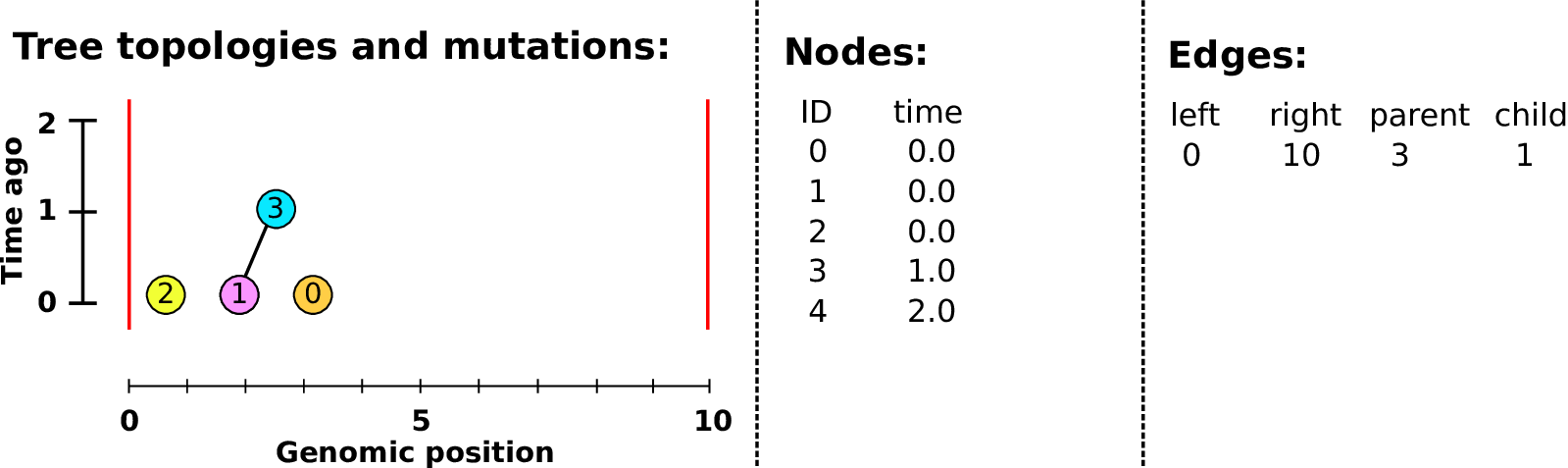
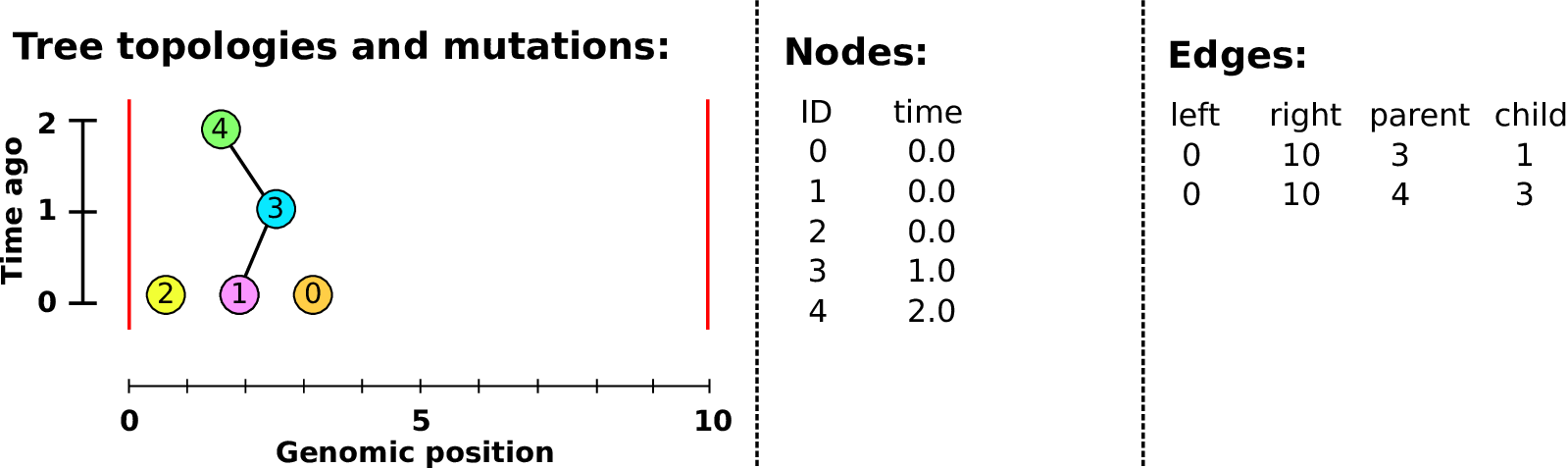
Nodes and edges
- Edges
-
Who inherits from who.
Records: interval (left, right); parent node; child node.
- Nodes
-
The ancestors those happen in.
Records: time ago (of birth); ID (implicit).
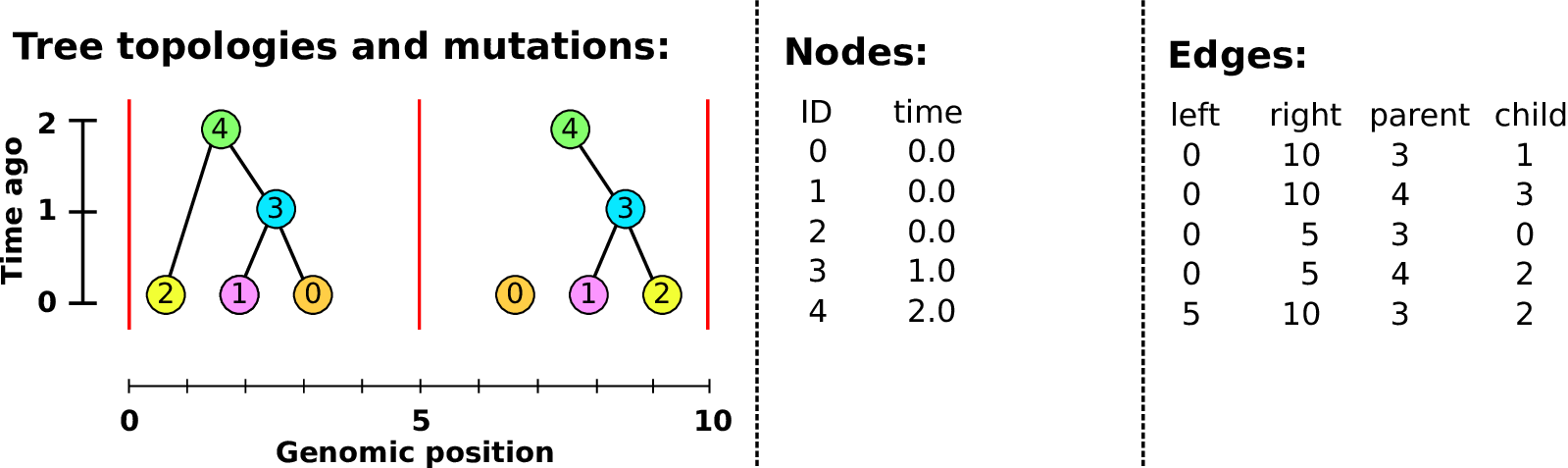
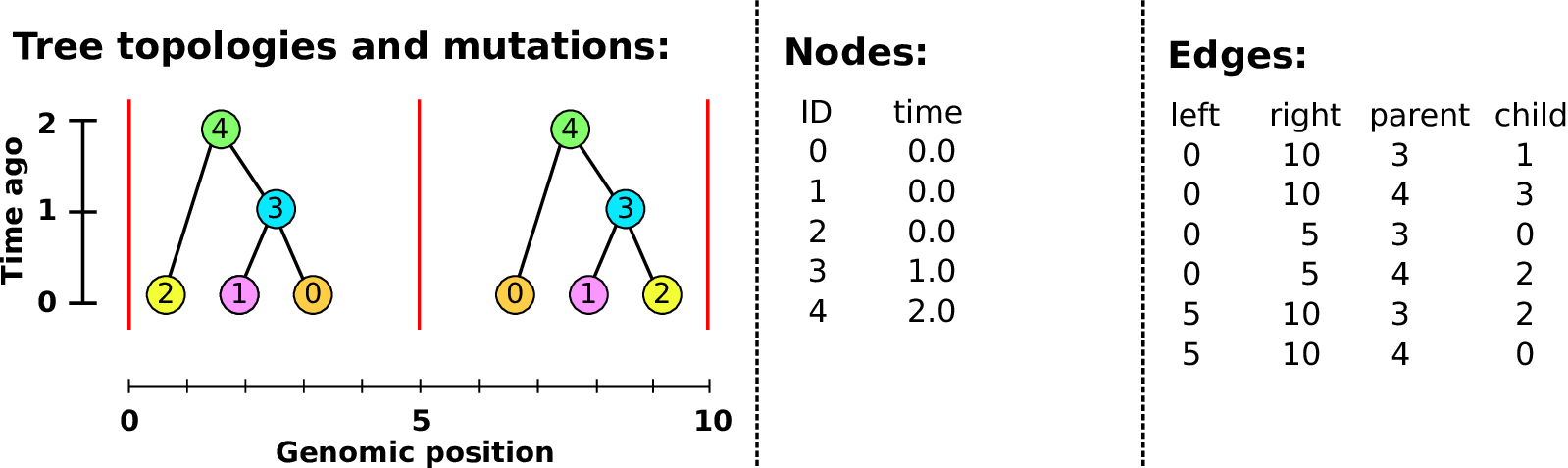
Sites and mutations
- Mutations
-
When state changes along the tree.
Records: site it occured at; node it occurred in; derived state.
- Sites
-
Where mutations fall on the genome.
Records: genomic position; ancestral (root) state; ID (implicit).
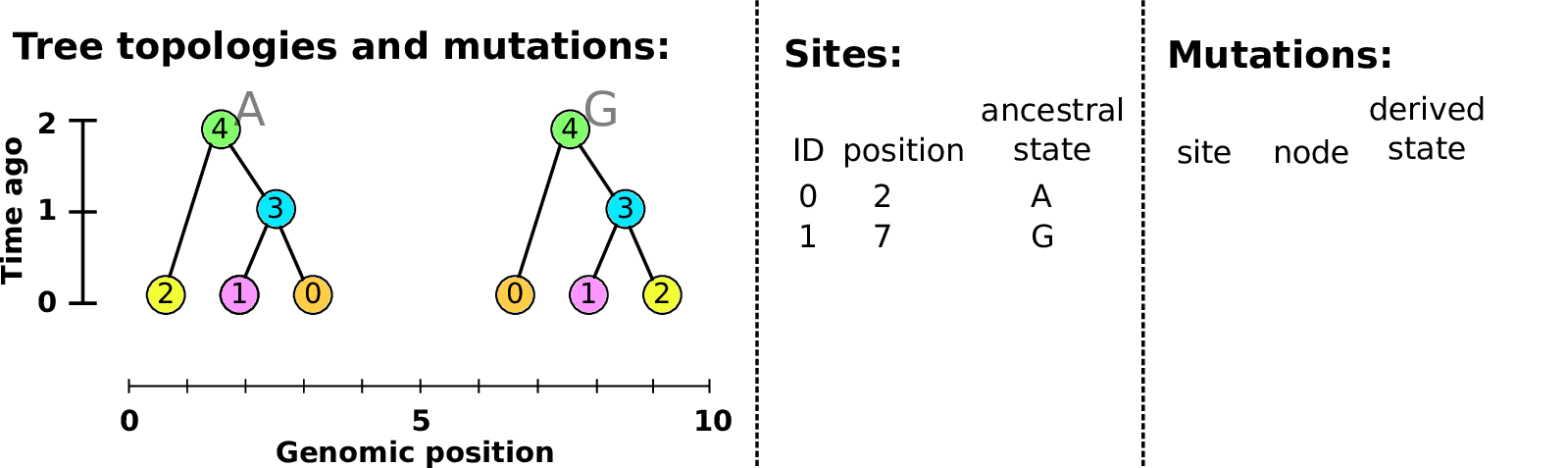
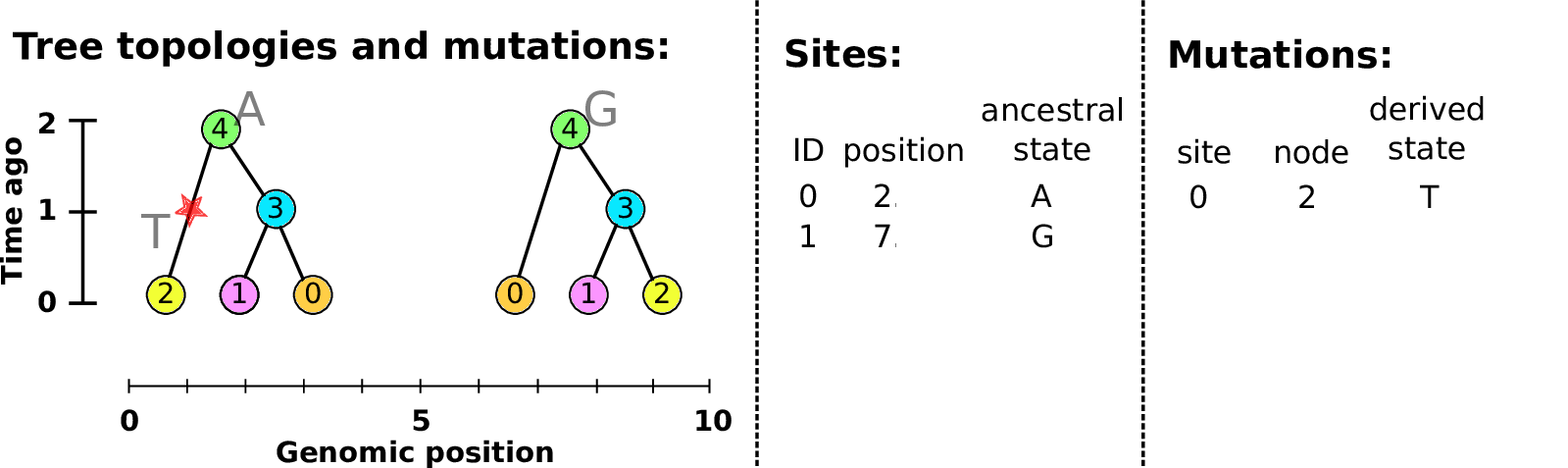
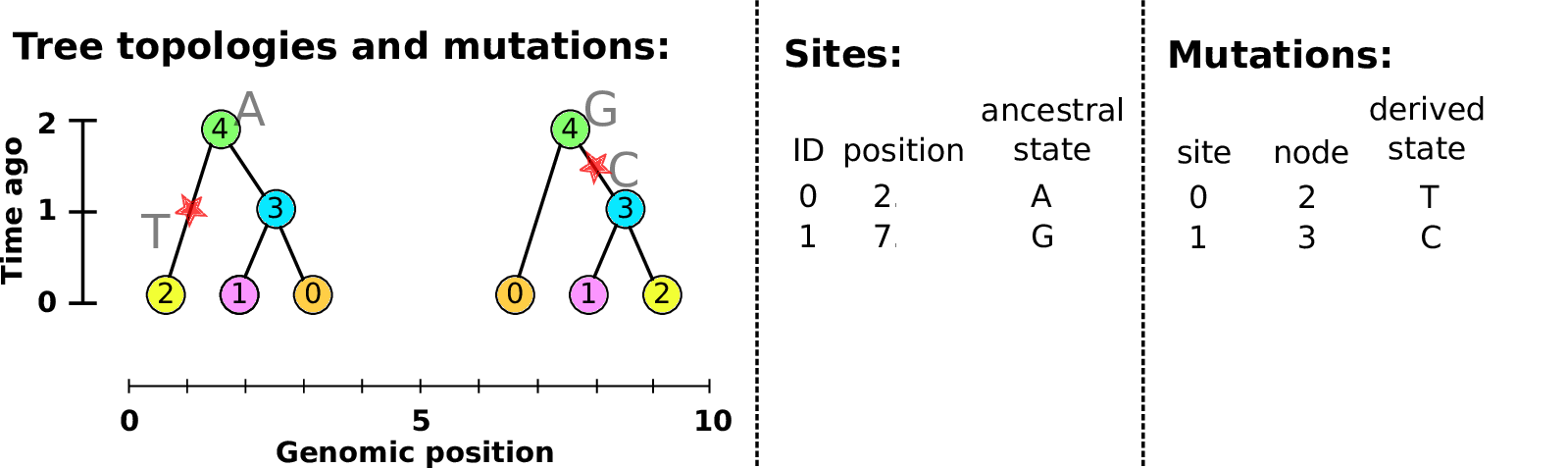
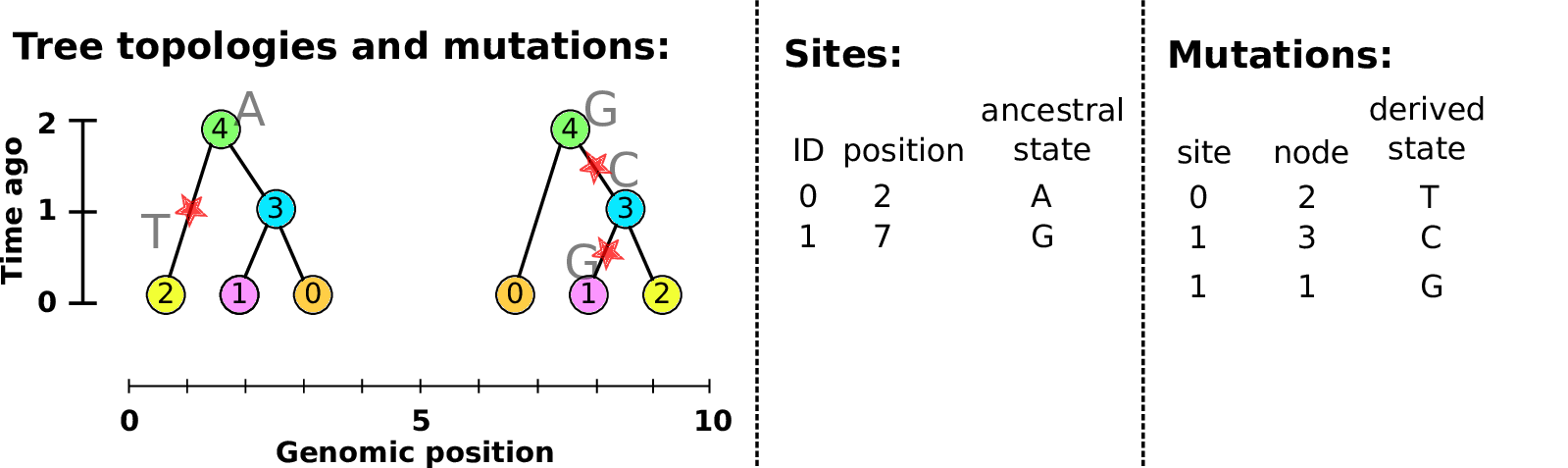
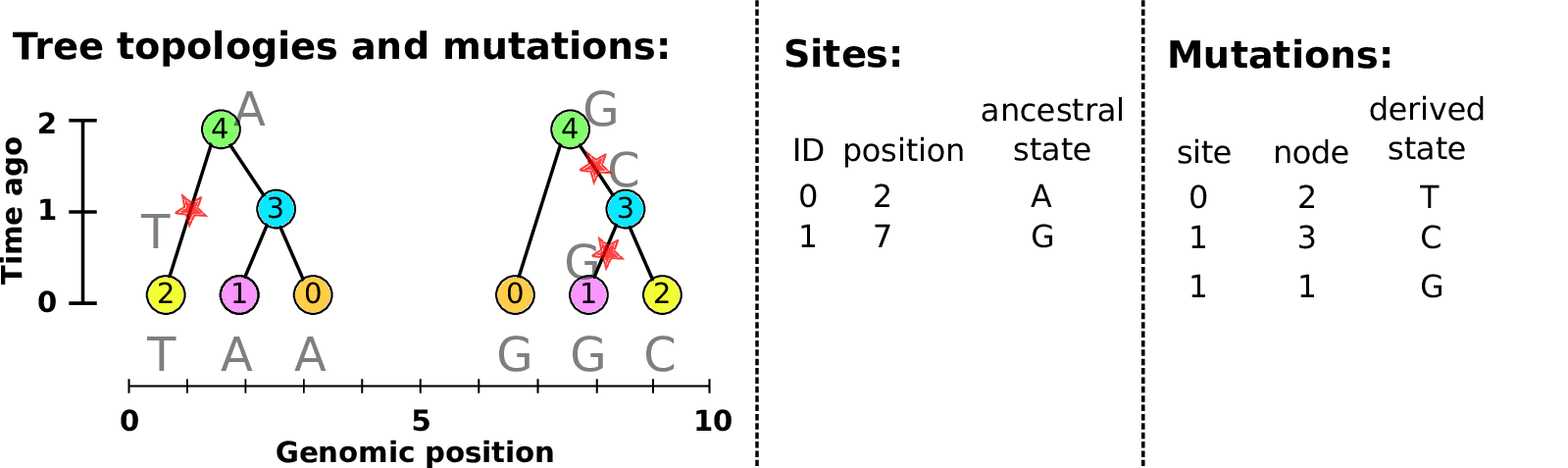
The result: an encoding of the genomes and all the genealogical trees.

How’s it work?
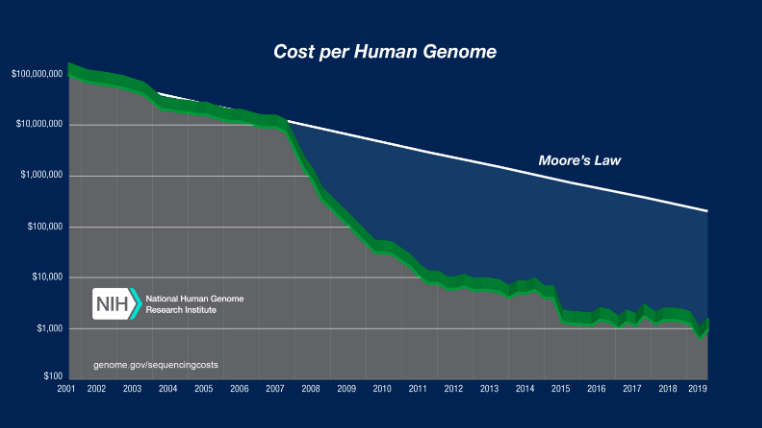
File sizes
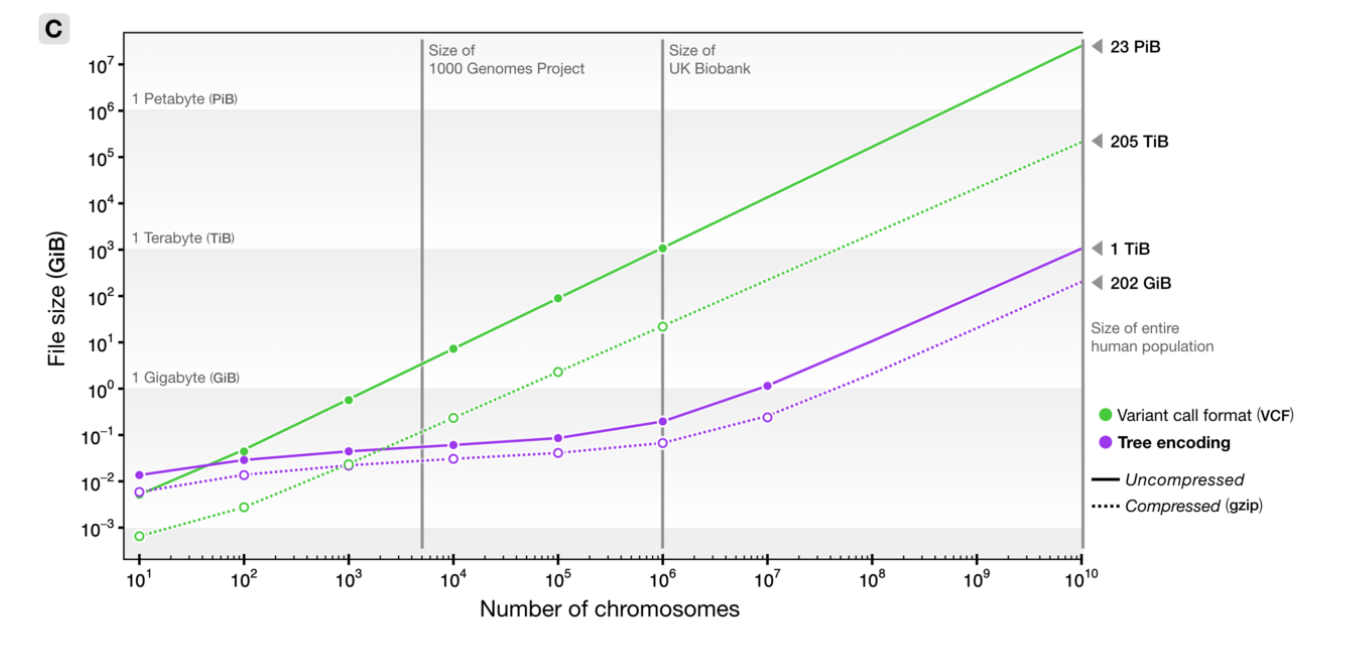
100Mb chromosomes; from Kelleher et al 2018, Inferring whole-genome histories in large population datasets, Nature Genetics
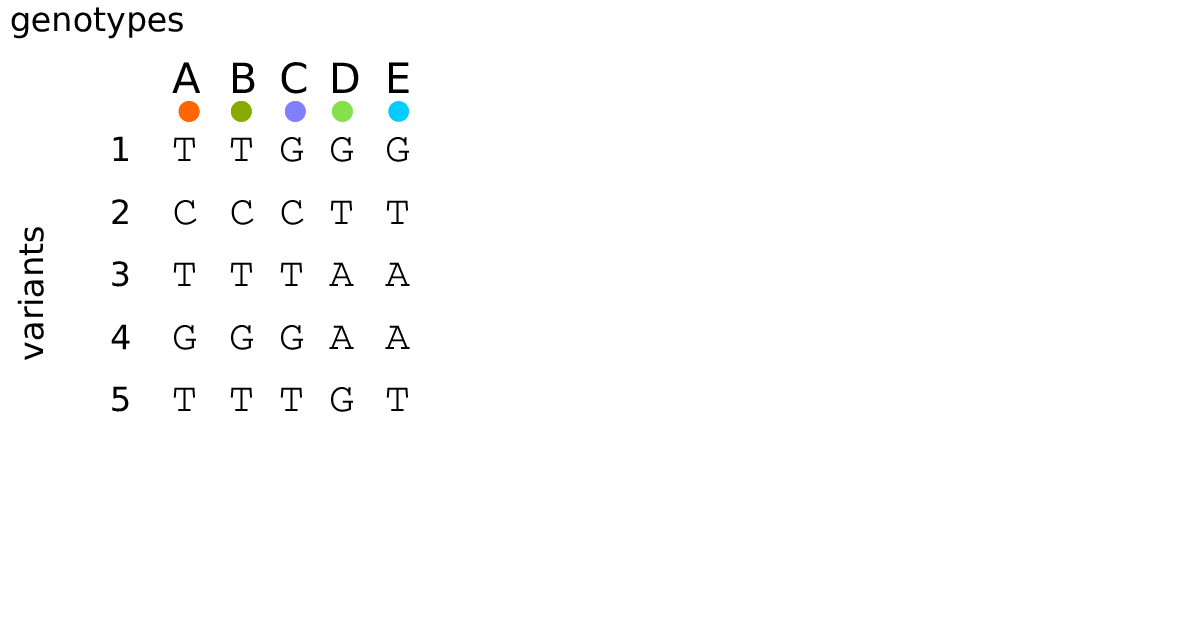
For \(N\) samples genotyped at \(M\) sites
Genotype matrix: \(O(NM)\)
\(N \times M\) things.
Tree sequence: \(O(N + T + M)\)
- \(2N-2\) edges for the first tree
- \(\sim 4\) edges per each of \(T\) trees
- \(M\) mutations

Fast genotype statistics

In SLiM
The main idea
If we record the tree sequence that relates everyone to everyone else,
after the simulation is over we can put neutral mutations down on the trees.
Since neutral mutations don’t affect demography,
this is equivalent to having kept track of them throughout.
This means recording the entire genetic history of everyone in the population, ever.
It is not clear this is a good idea.
Tree recording strategy
Every time an individual is born, we must:
- add each gamete to the Node Table,
- add entries to the Edge Table recording which parent each gamete inherited each bit of genome from, and
- add any new selected mutations to the Mutation Table and (if necessary) their locations to the Site Table.

This produces waaaaay too much data.
We won’t end up needing the entire history of everyone ever,
but we won’t know what we’ll need until later.
How do we get rid of the extra stuff?
Simplification.
Wrap up
Software
Everything is efficient, open, and tested.

tskit: tree sequence toolsstdpopsim: a library of “standard” simulation toolsmsprime: coalescent simulator,SLiM: forwards evolutionary simulator