Landscapes in population genetics:
ecology, evolution, conservation, and simulation
Peter Ralph
Program in Ecology, Evolution & Conservation Biology
UIUC // 3 February 2021
Outline
Outline of the talk
- Big picture
- Tools
- Applications
Adaptation, and genetic variation
G6PD deficiency allele frequencies
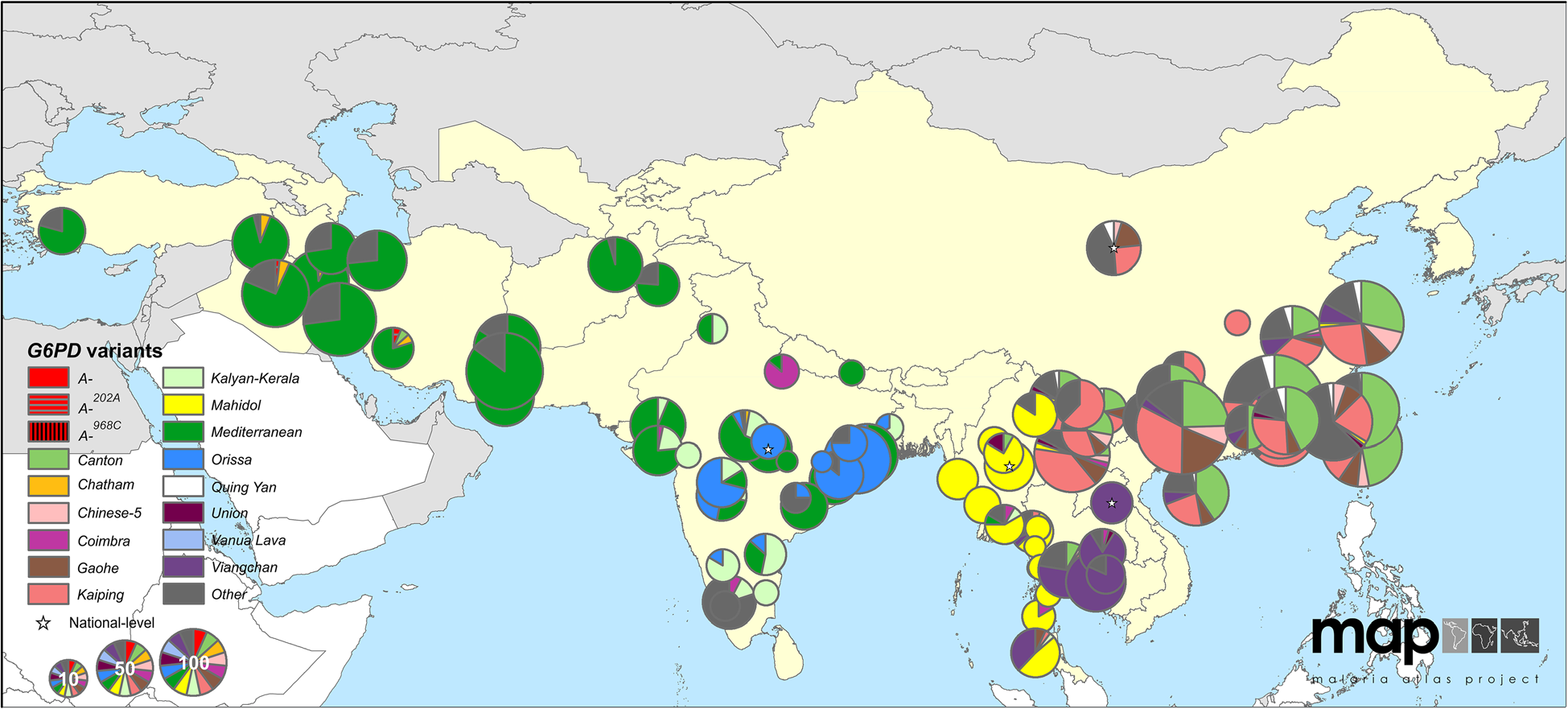
Human G6PD variants (Howes et al 2013)
- over 130 G6PD deficiency alleles; 34 variants at high frequency
- provide protection against malaria but increase risk of anemia
- Estimated ages 40-400 generations

- Dark-pigmented mammals and reptiles on volcanic outcrops in the Southwest. (Dice, Benson 1936)
- ‘Dark’ allele beneficial on outcrops, deleterious elsewhere.
- MC1R: basis is shared between species but not between populations (Nachman, Hoekstra)
“isolation by distance”
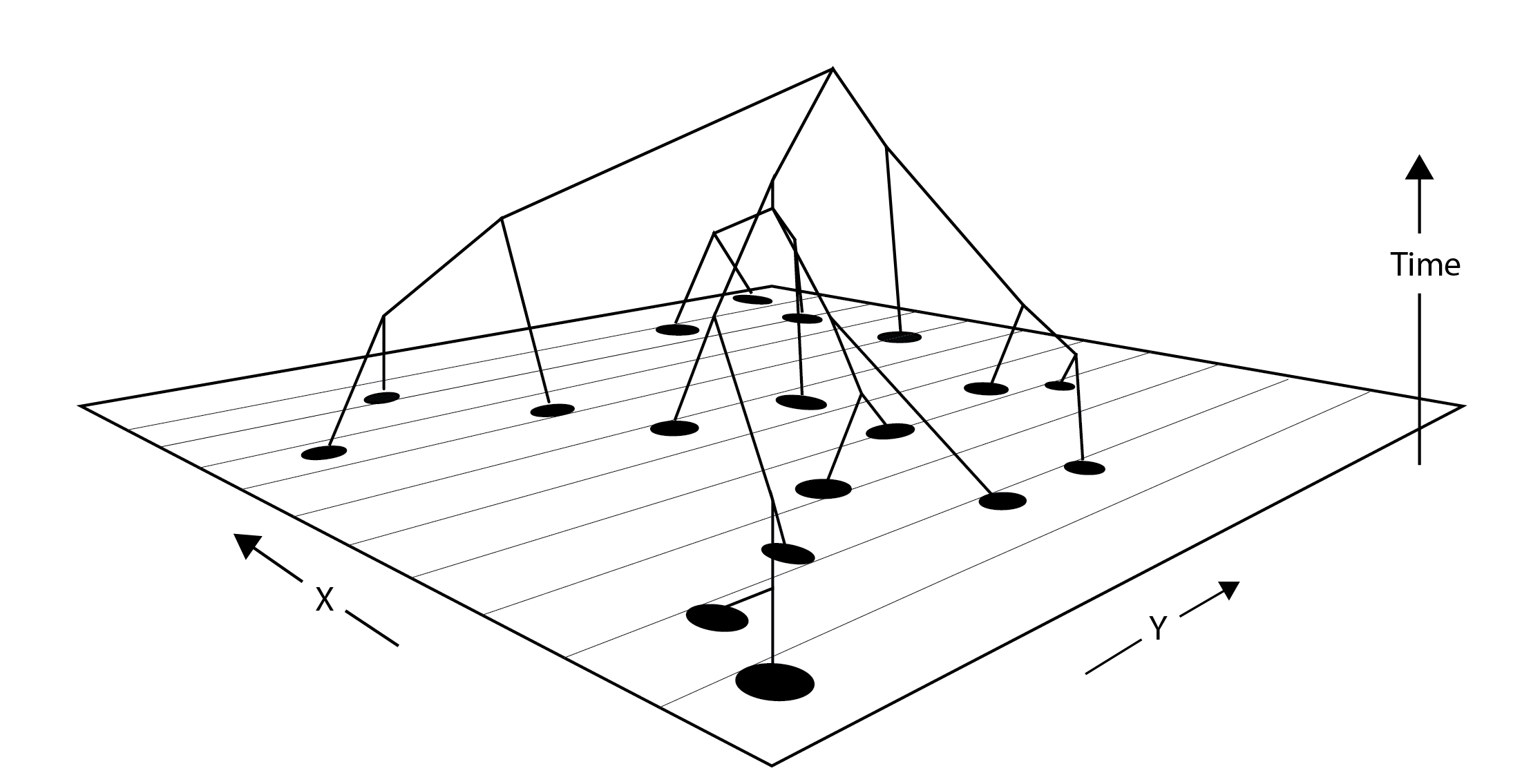
by CJ Battey
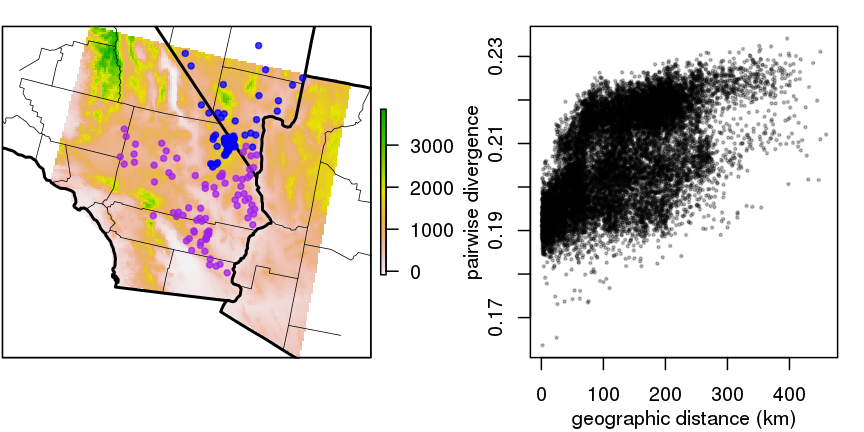
- genetic versus geographic distance between pairs of 272 desert tortoises (McCartney-Melstad, Shaffer)
- clouds are comparisons within/between the two colors
How much of the genome is under selection?
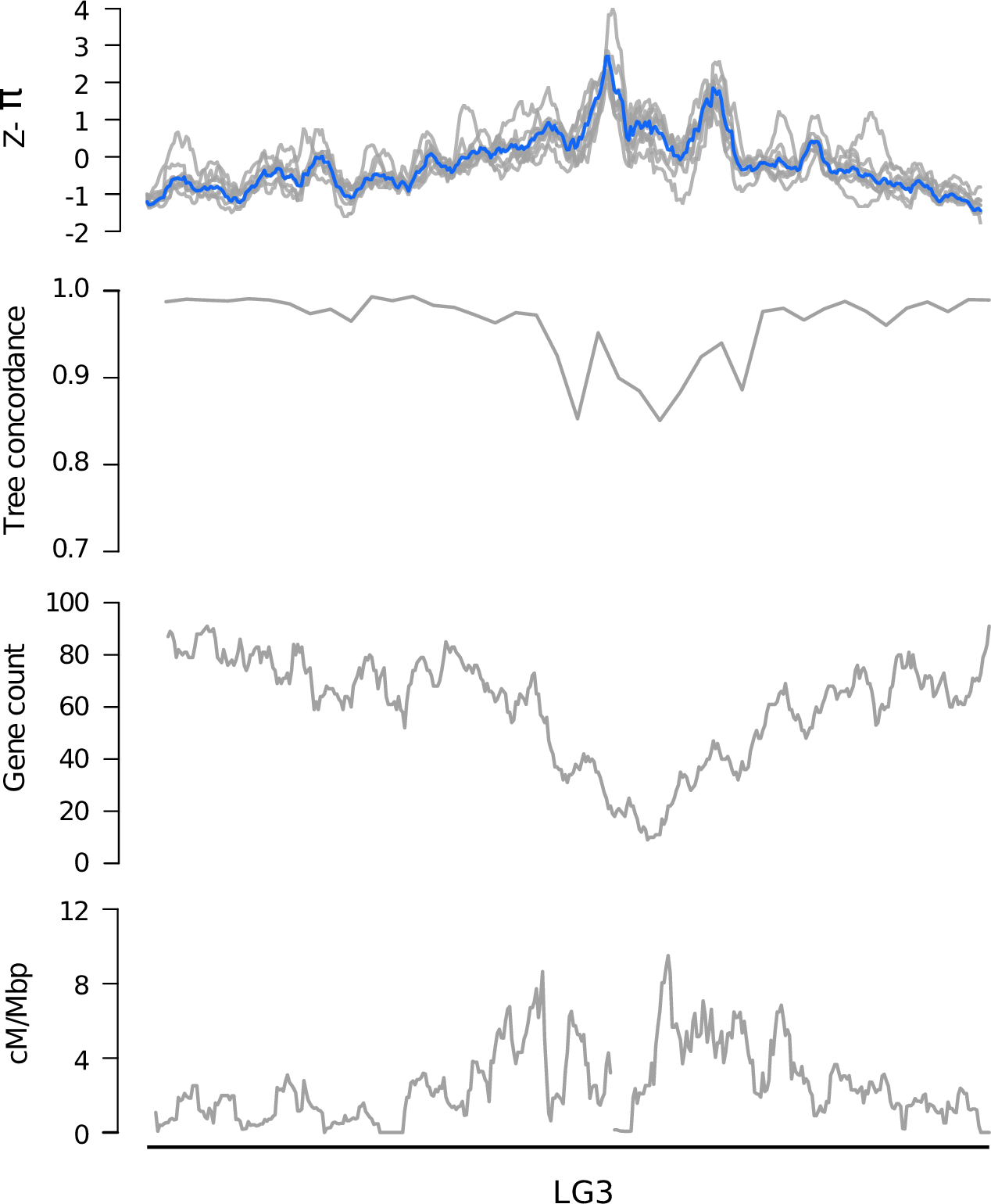
Genomic landscapes
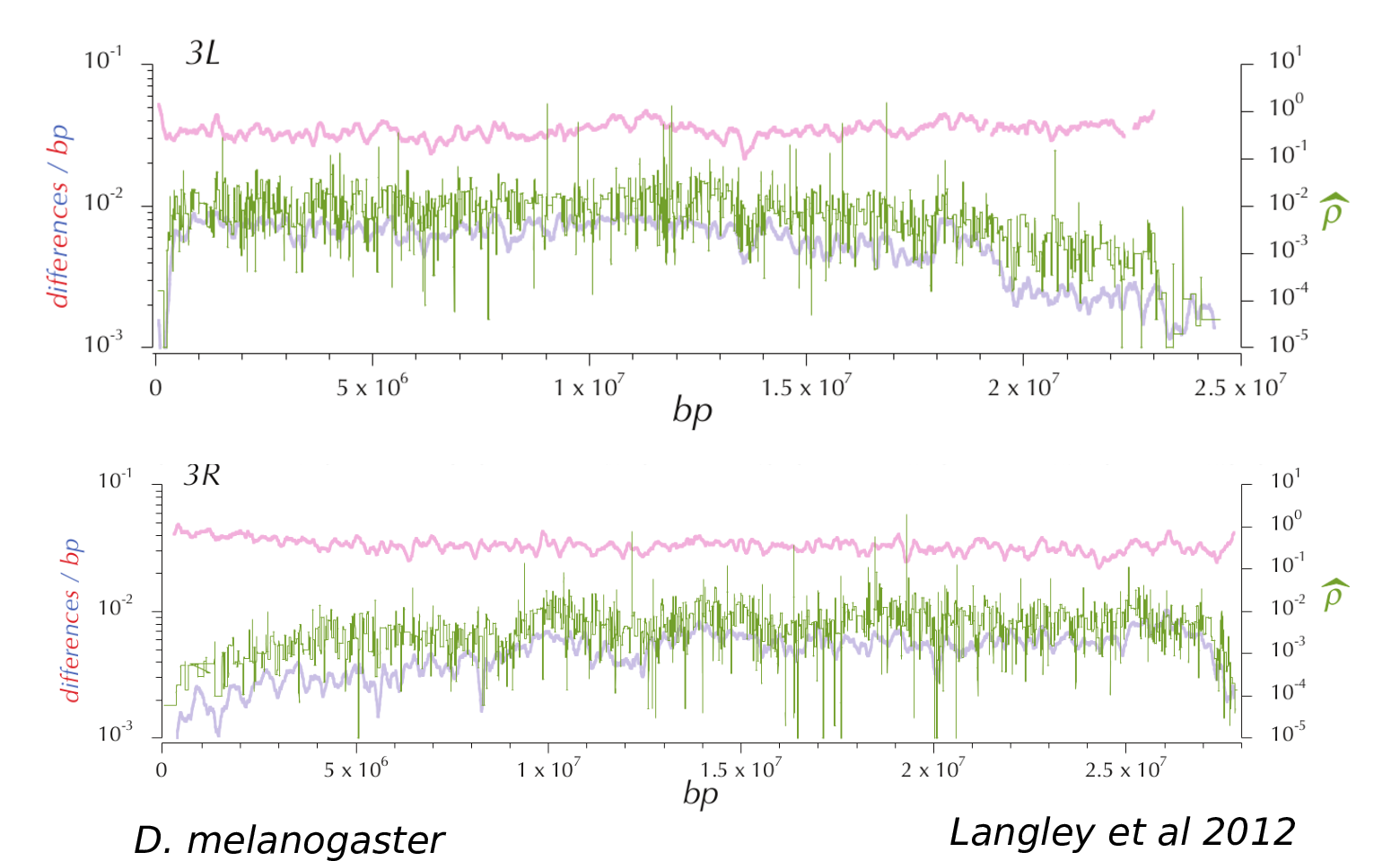
Diversity correlates with recombination rate
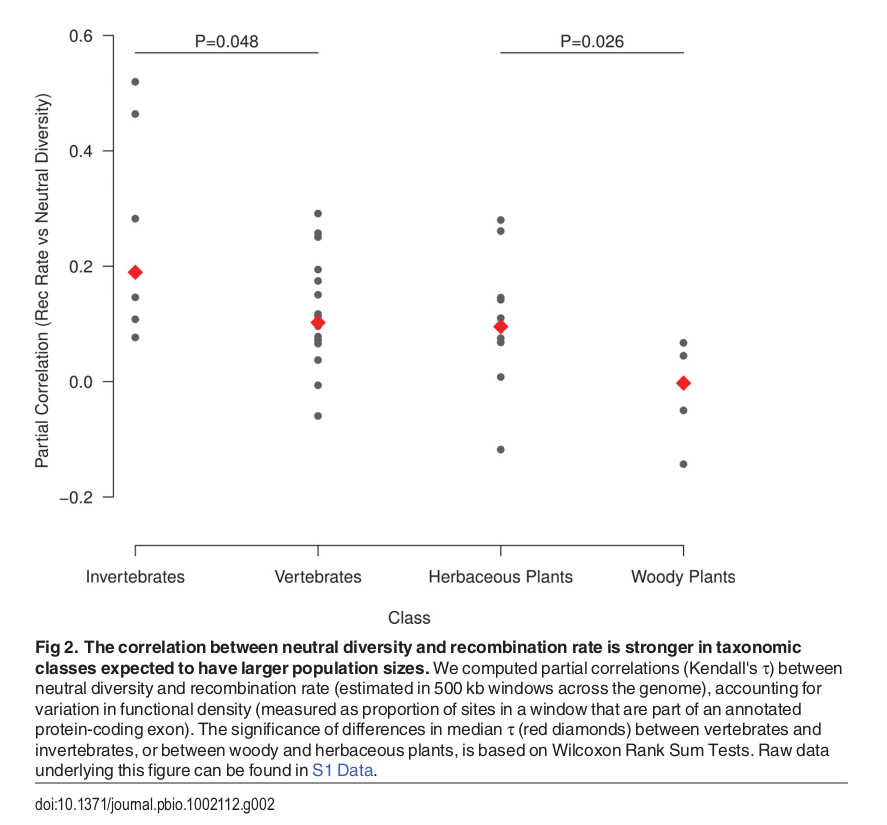
Hudson 1994; Cutter & Payseur 2013; Corbett-Detig et al 2015
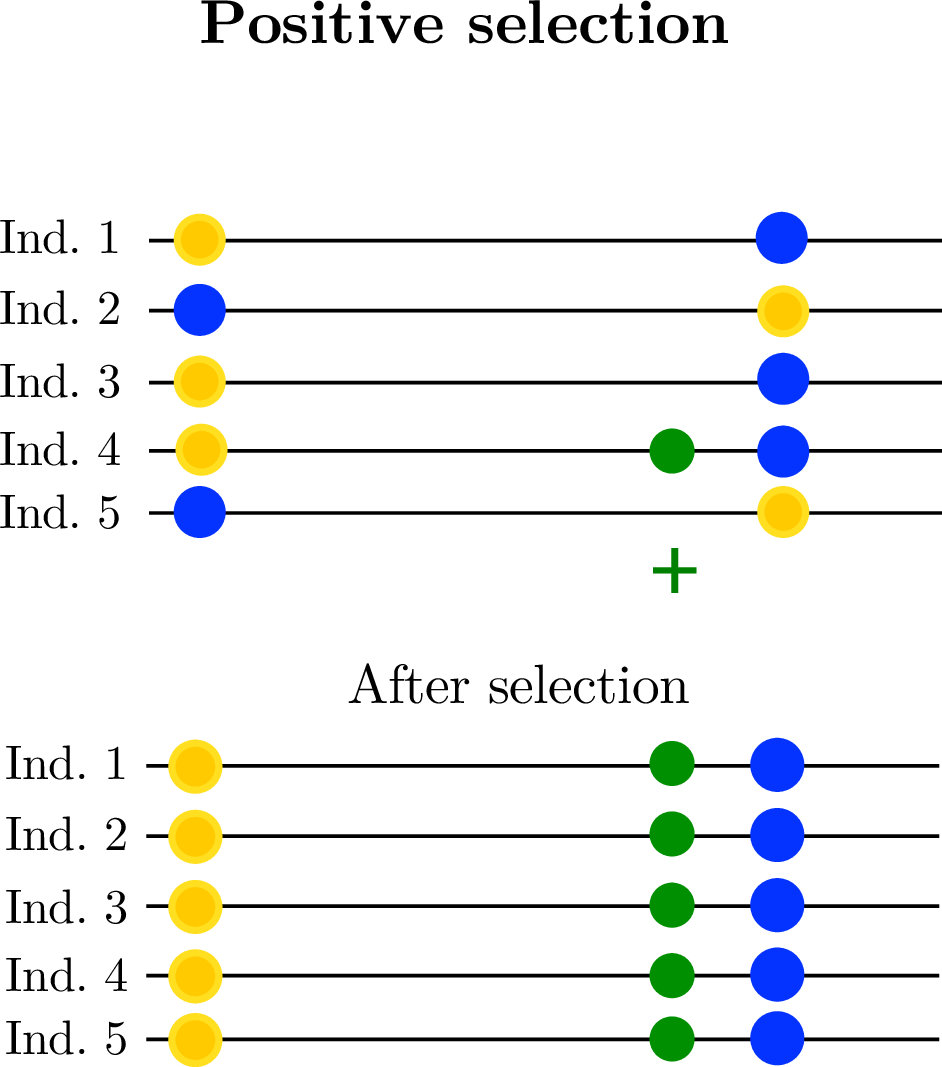
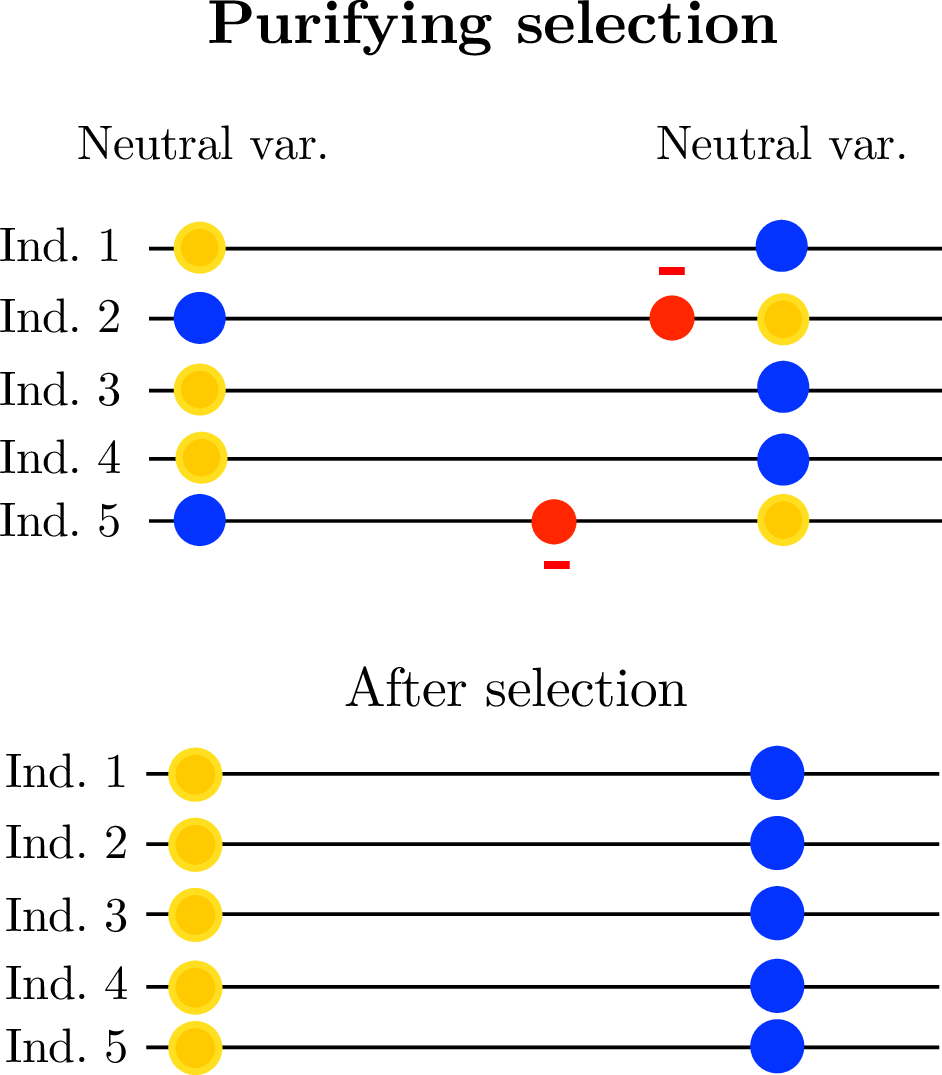
- linked selection
The indirect effects of selection on genomic locations that are linked to the sites under selection by a lack of recombination.
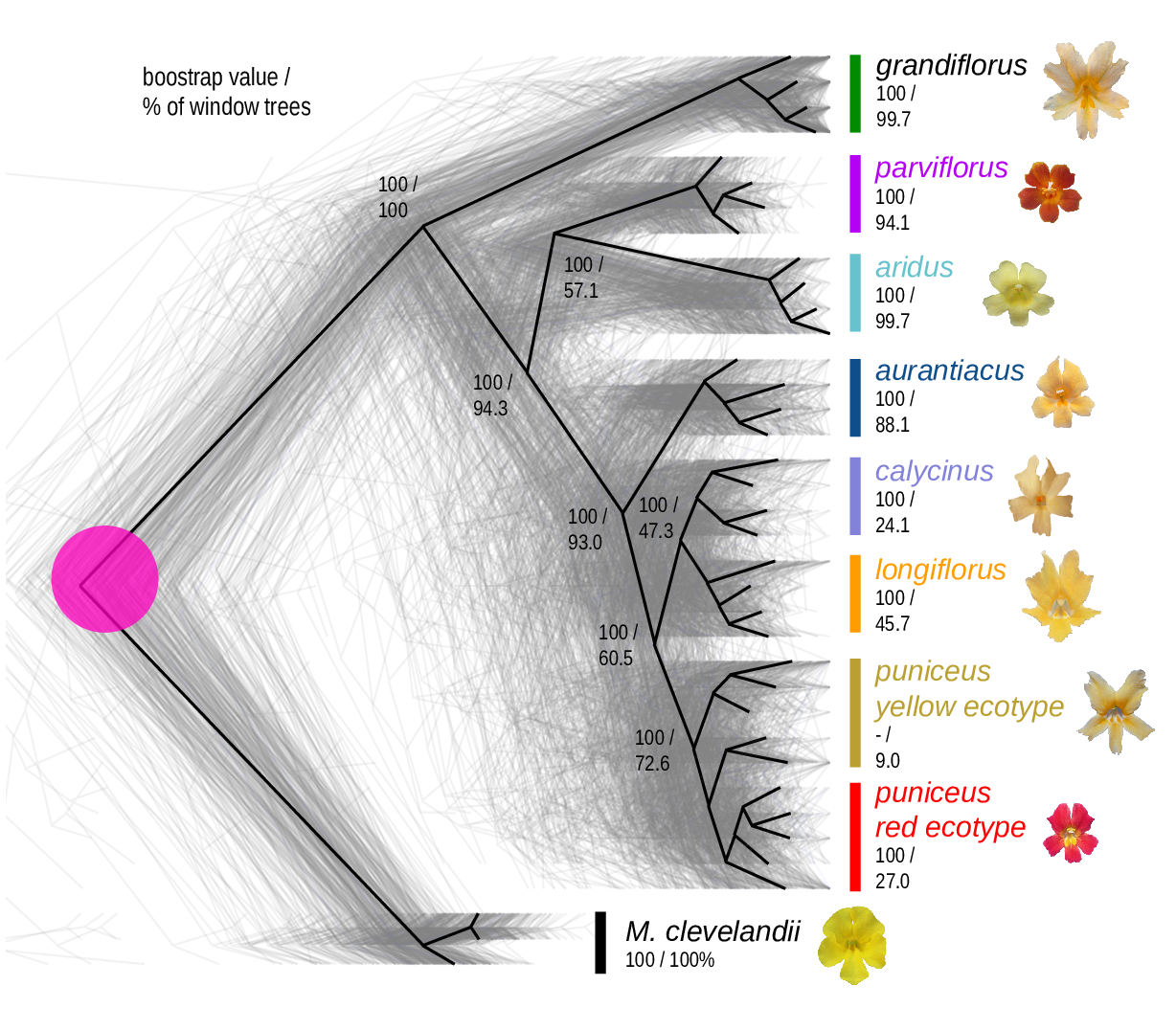
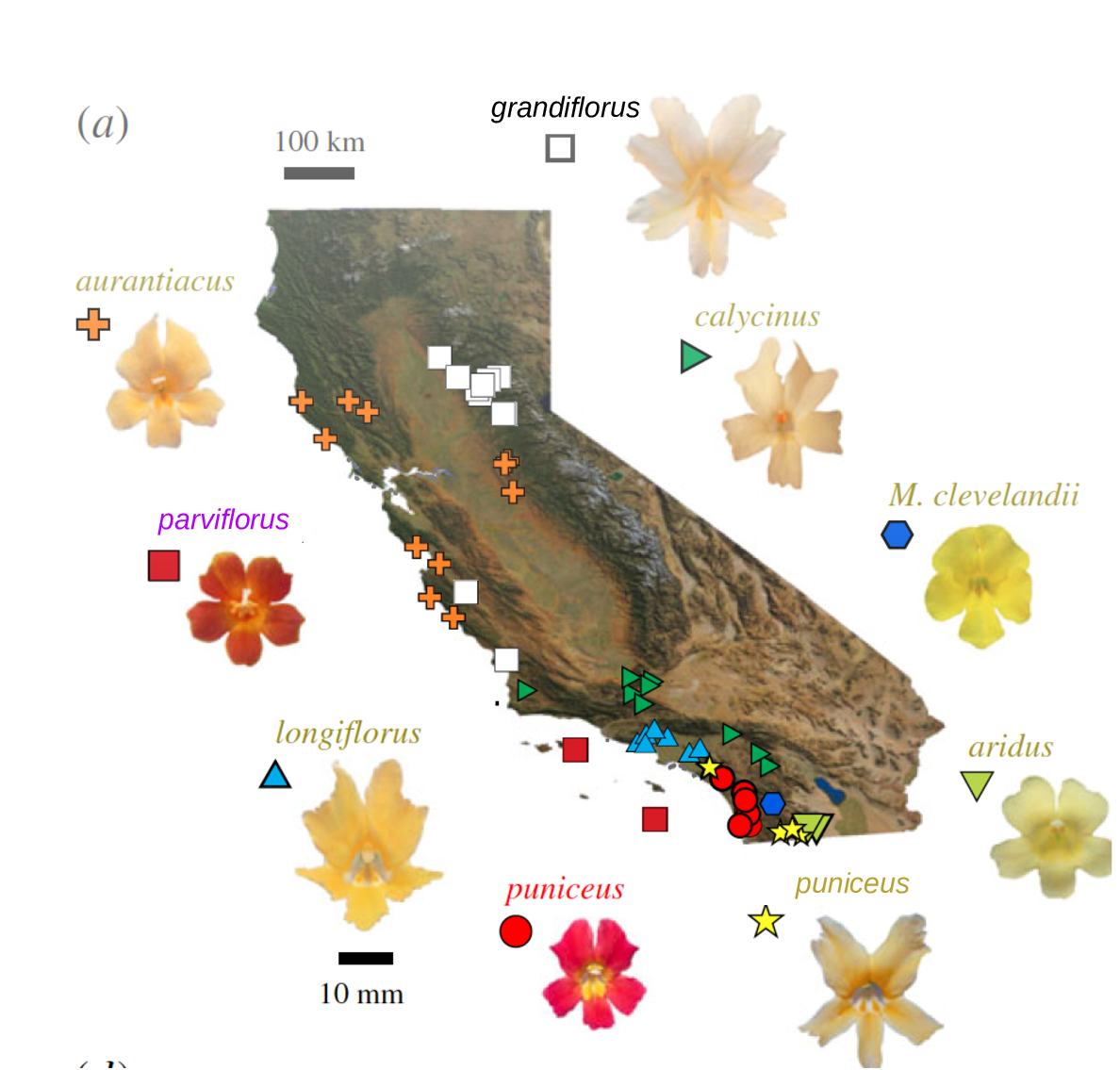
The Mimulus aurantiacus species complex
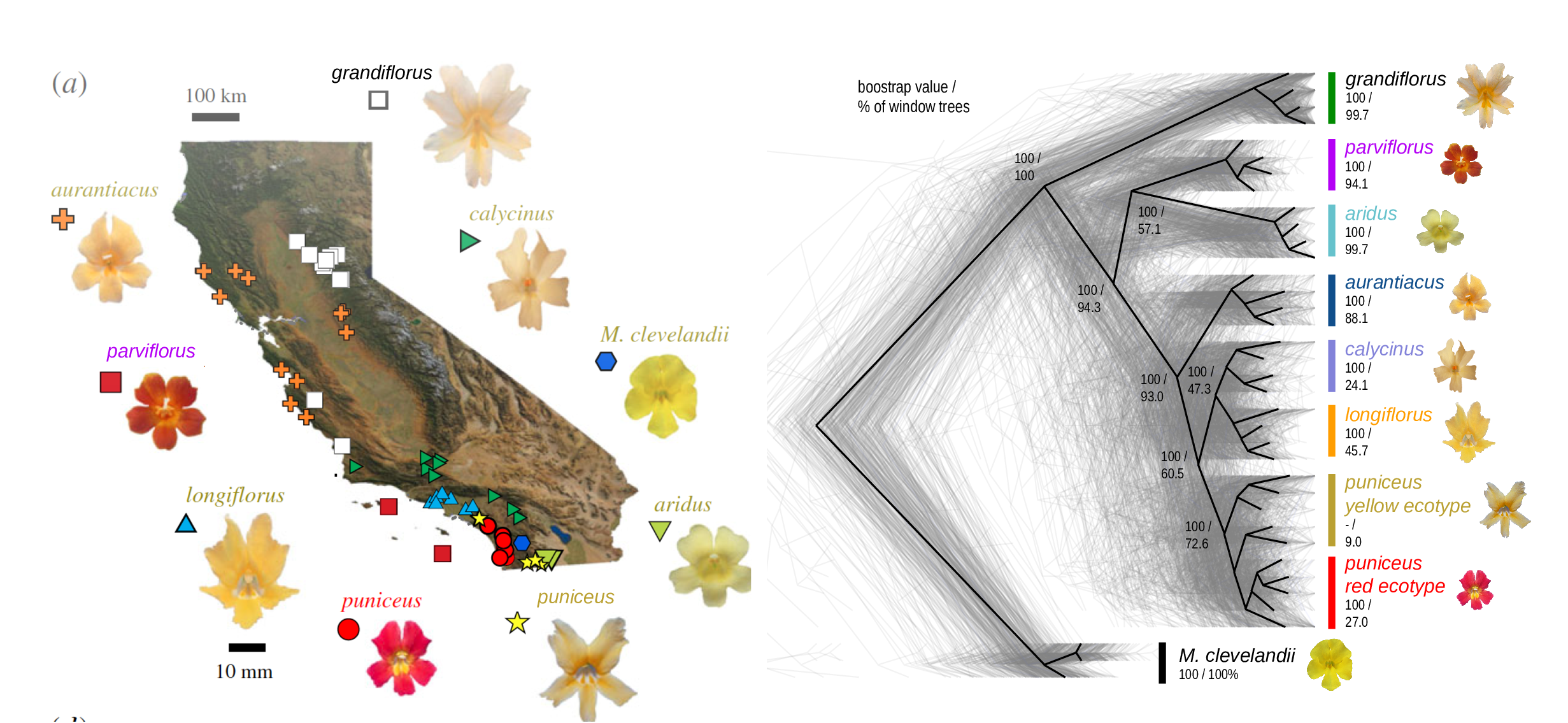
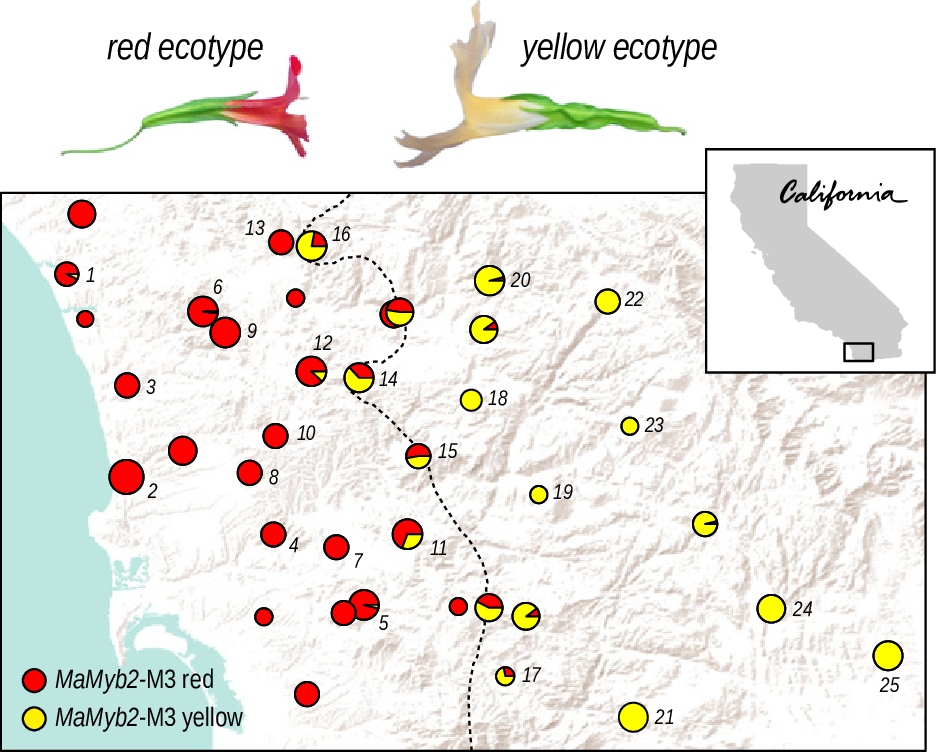
From Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers, Stankowski, Chase, Fuiten, Rodrigues, Ralph, and Streisfeld; PLoS Bio 2019.



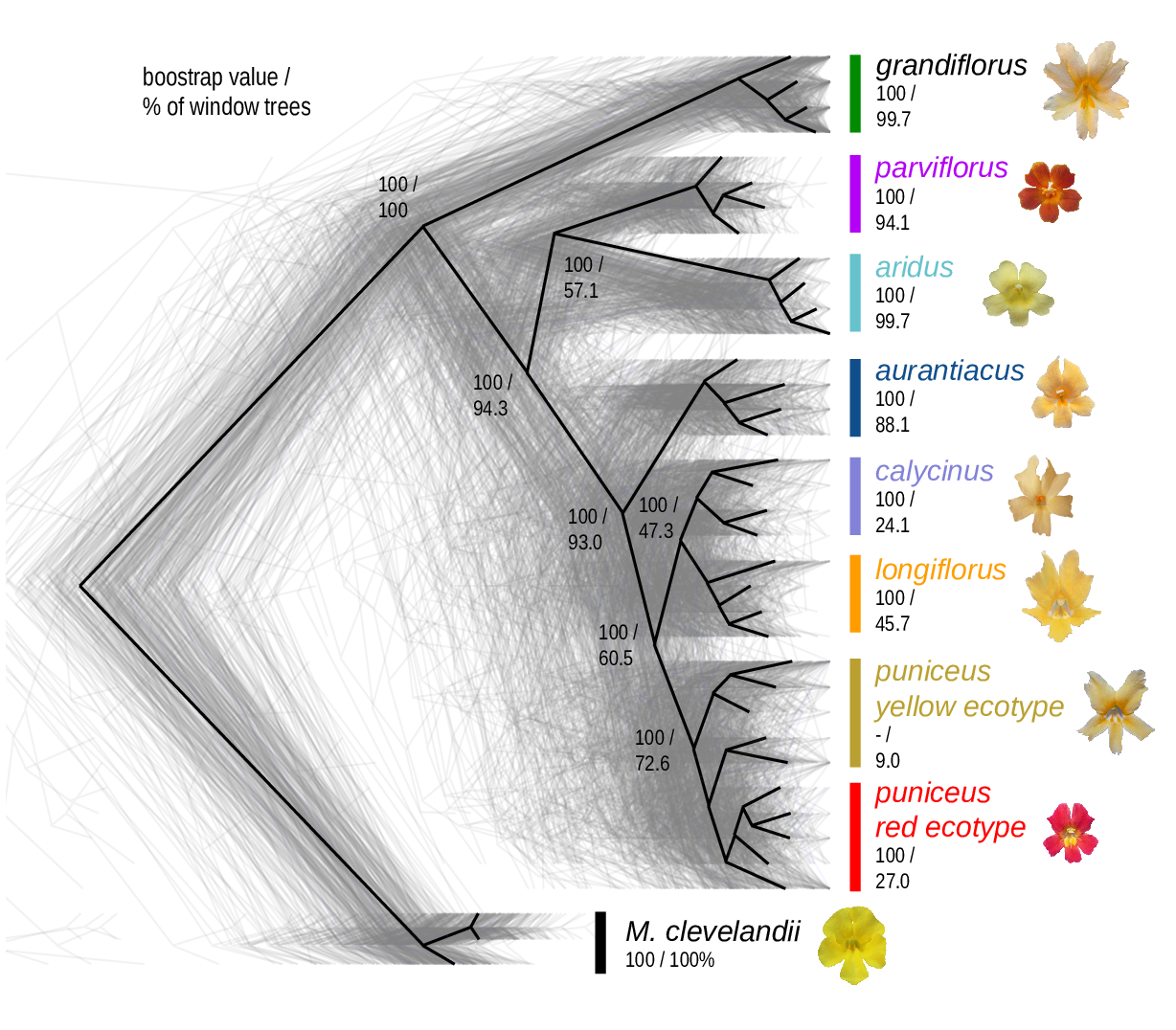
The data:
- chromosome-level genome assembly
- \(20\times\) coverage of 8 taxa and outgroup (M.clevelandii)
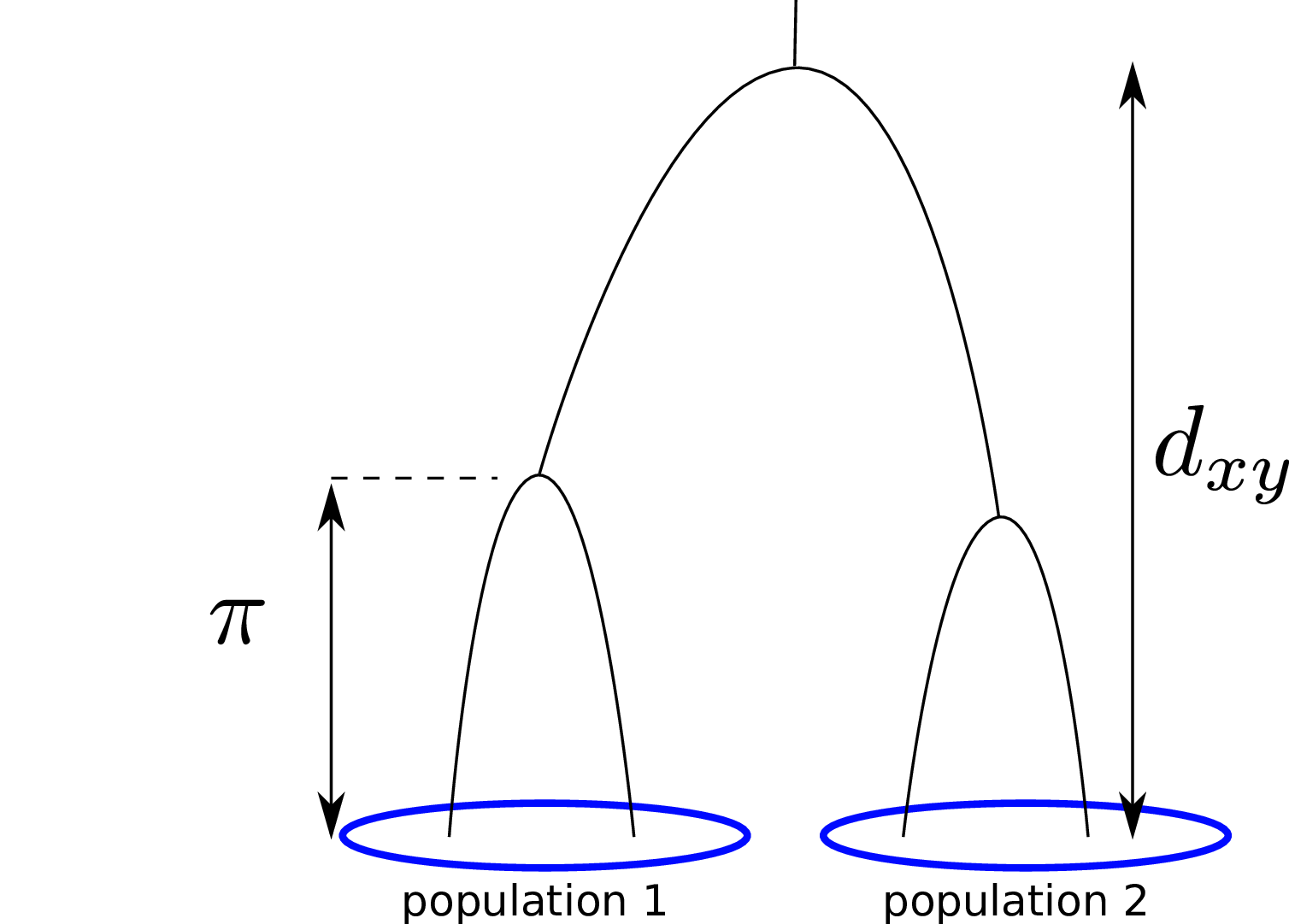
- diversity (\(\pi\)), divergence (\(d_{xy}\)), and differentiation (\(F_{ST}\)) in windows
- 36 pairwise comparisons among 9 taxa
- estimates of recombination rate and gene density from map and annotation
\[ \begin{aligned} \pi &= \text{ (within-pop genetic distance) } \\ d_{xy} &= \text{ (between-pop genetic distance) } \end{aligned} \]
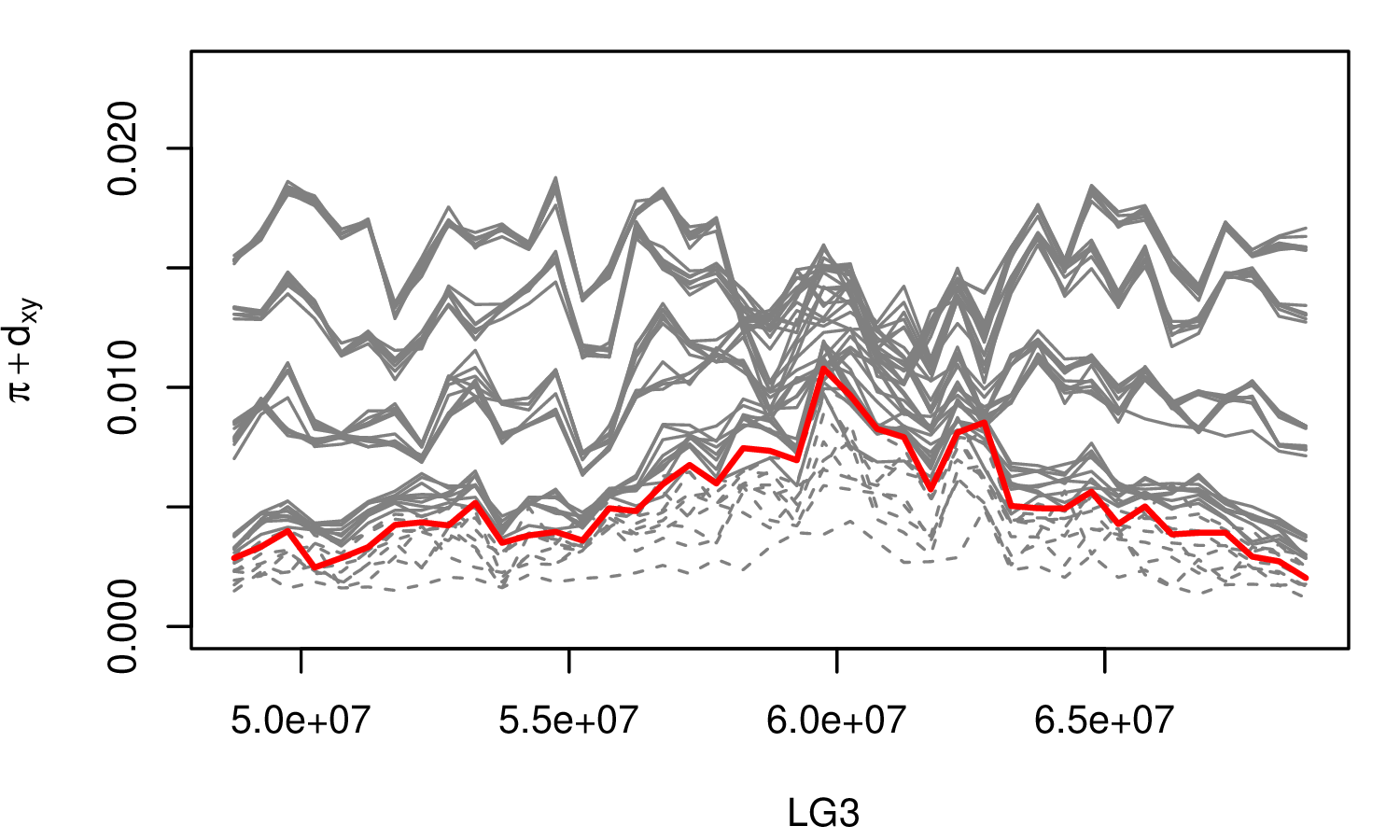
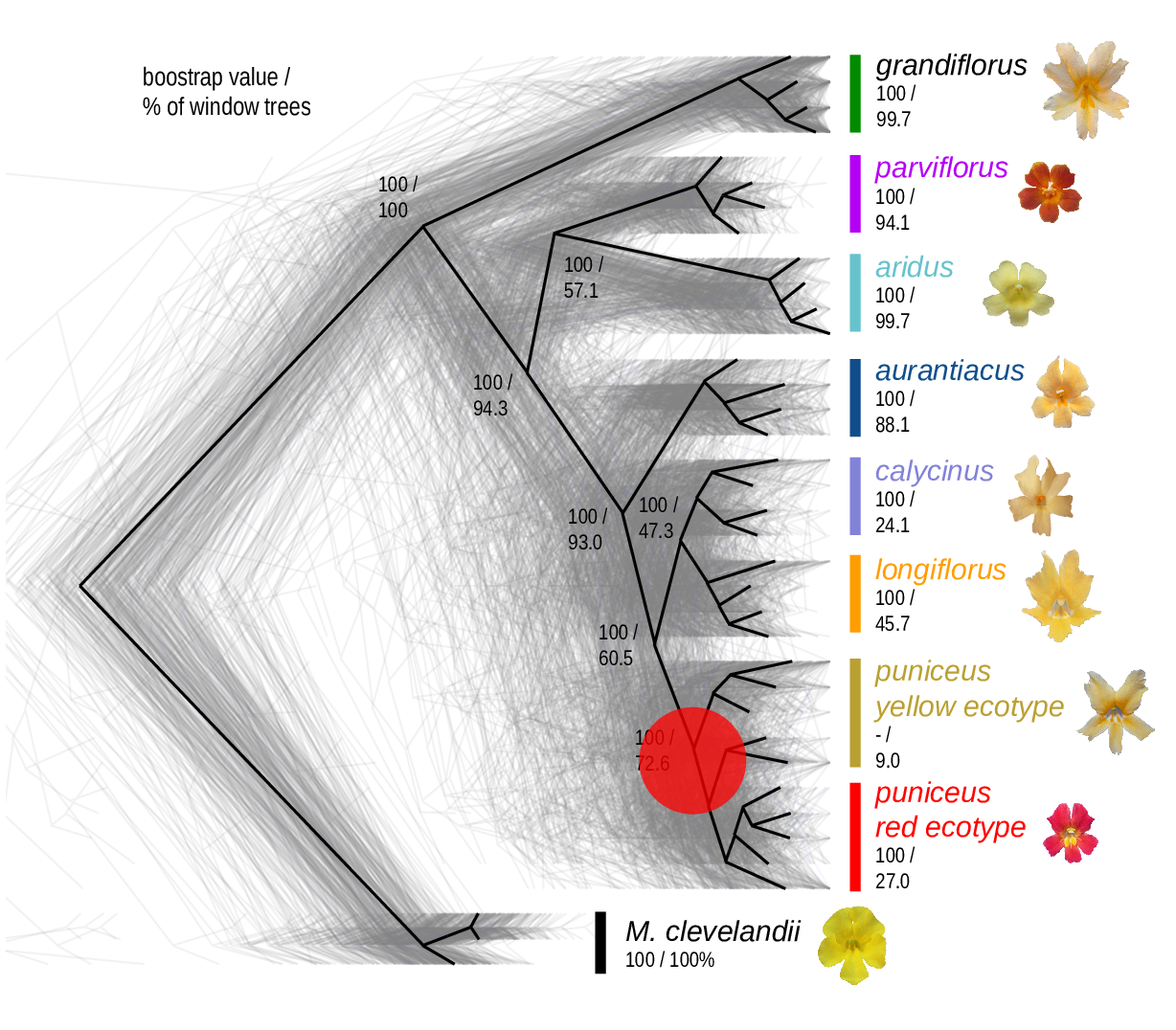
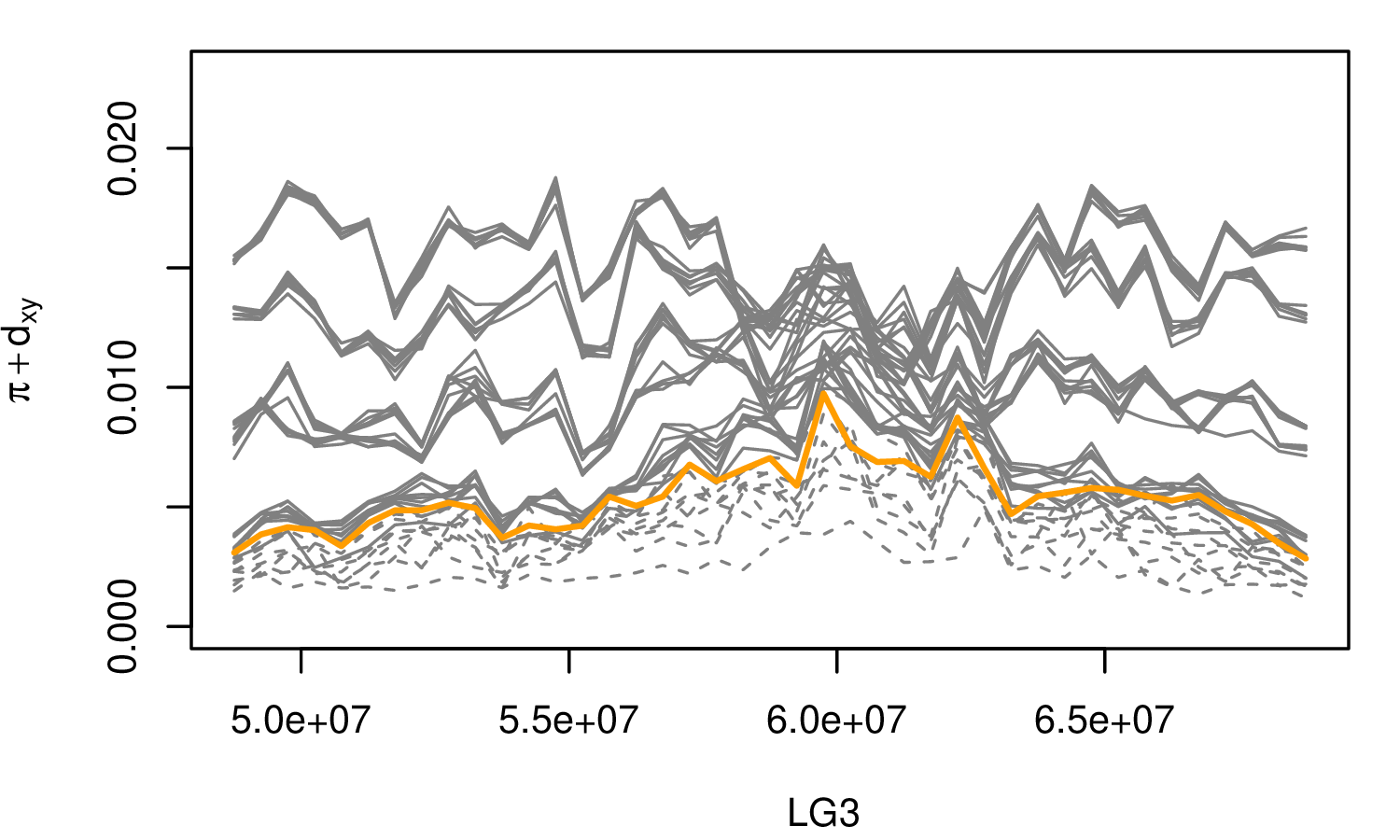
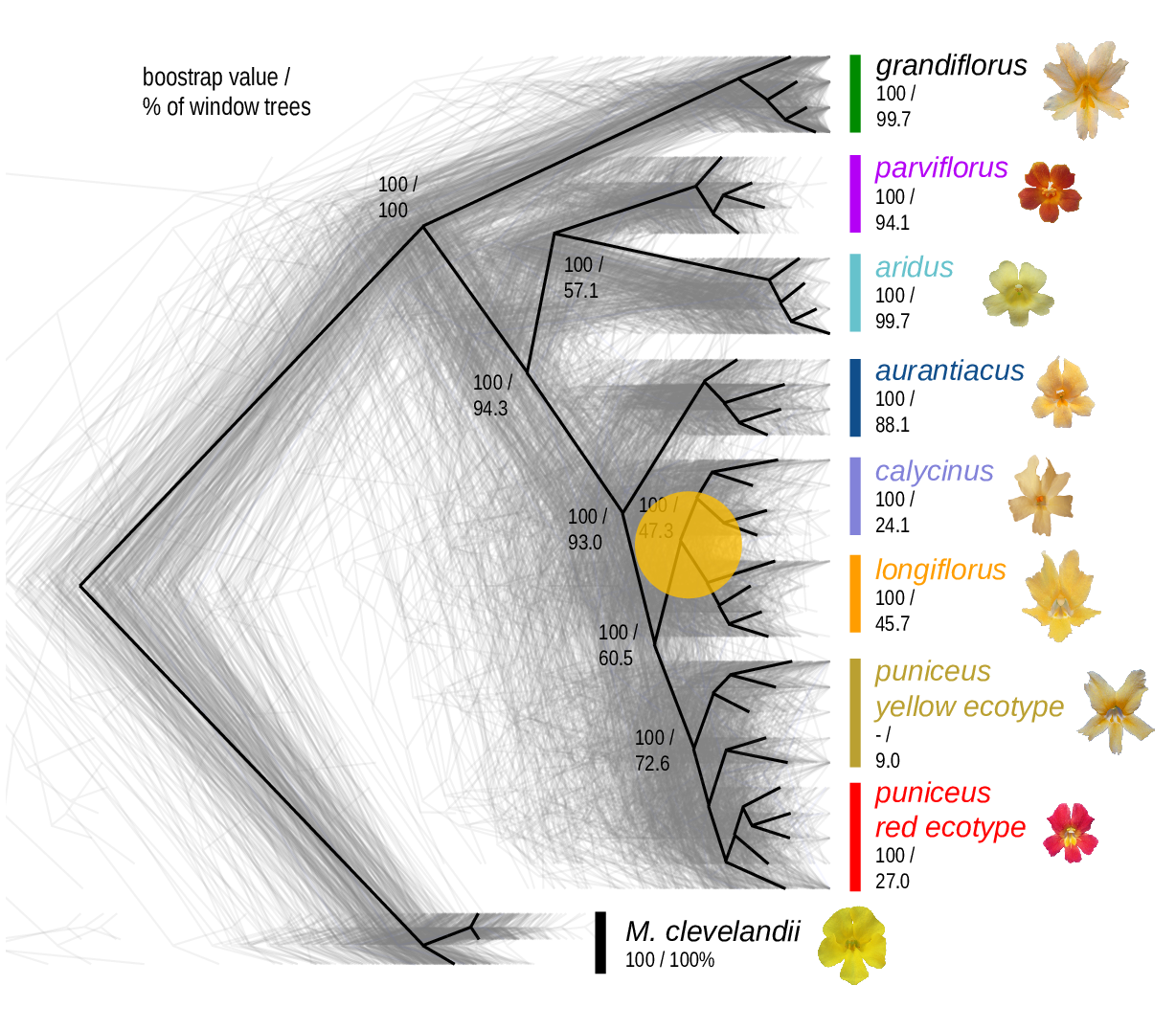
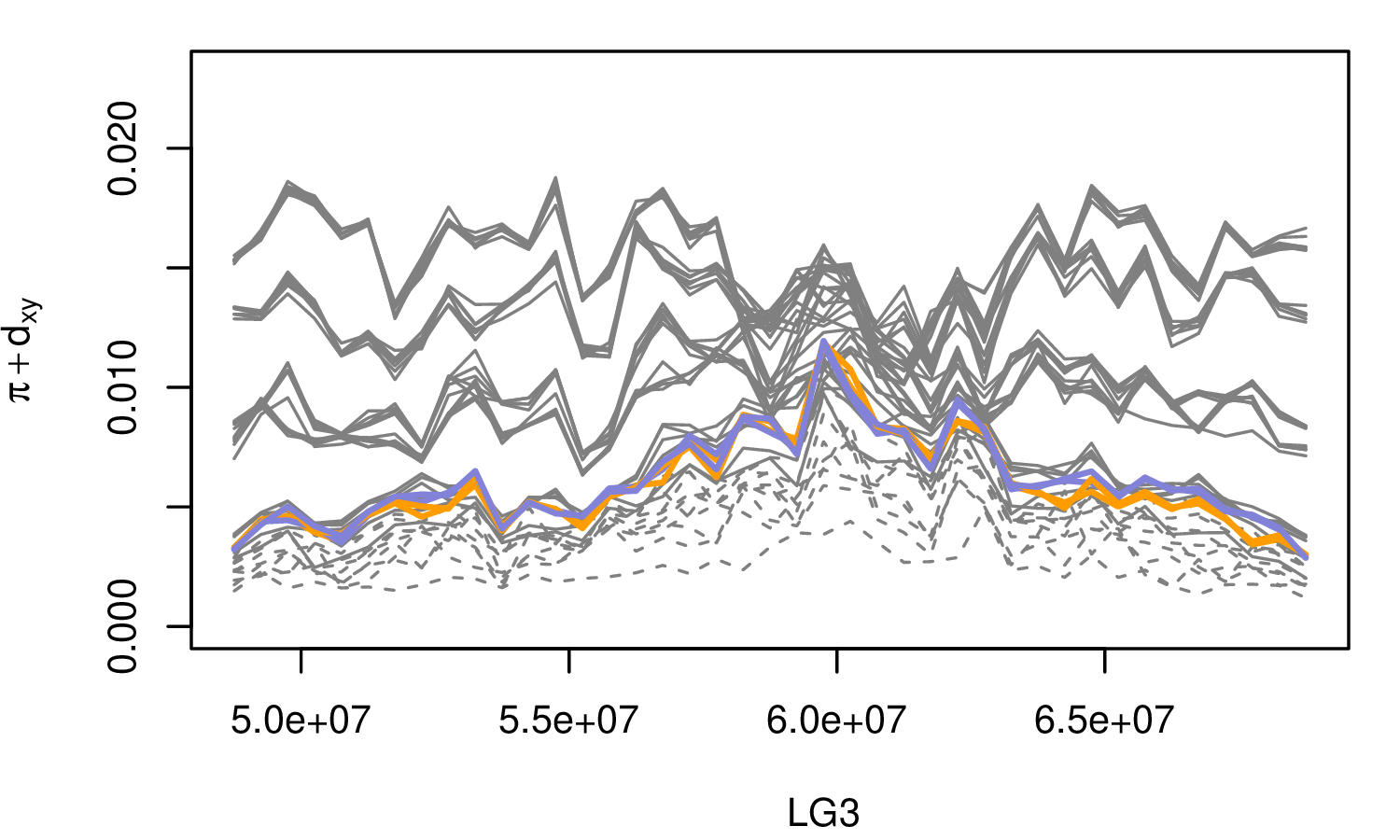
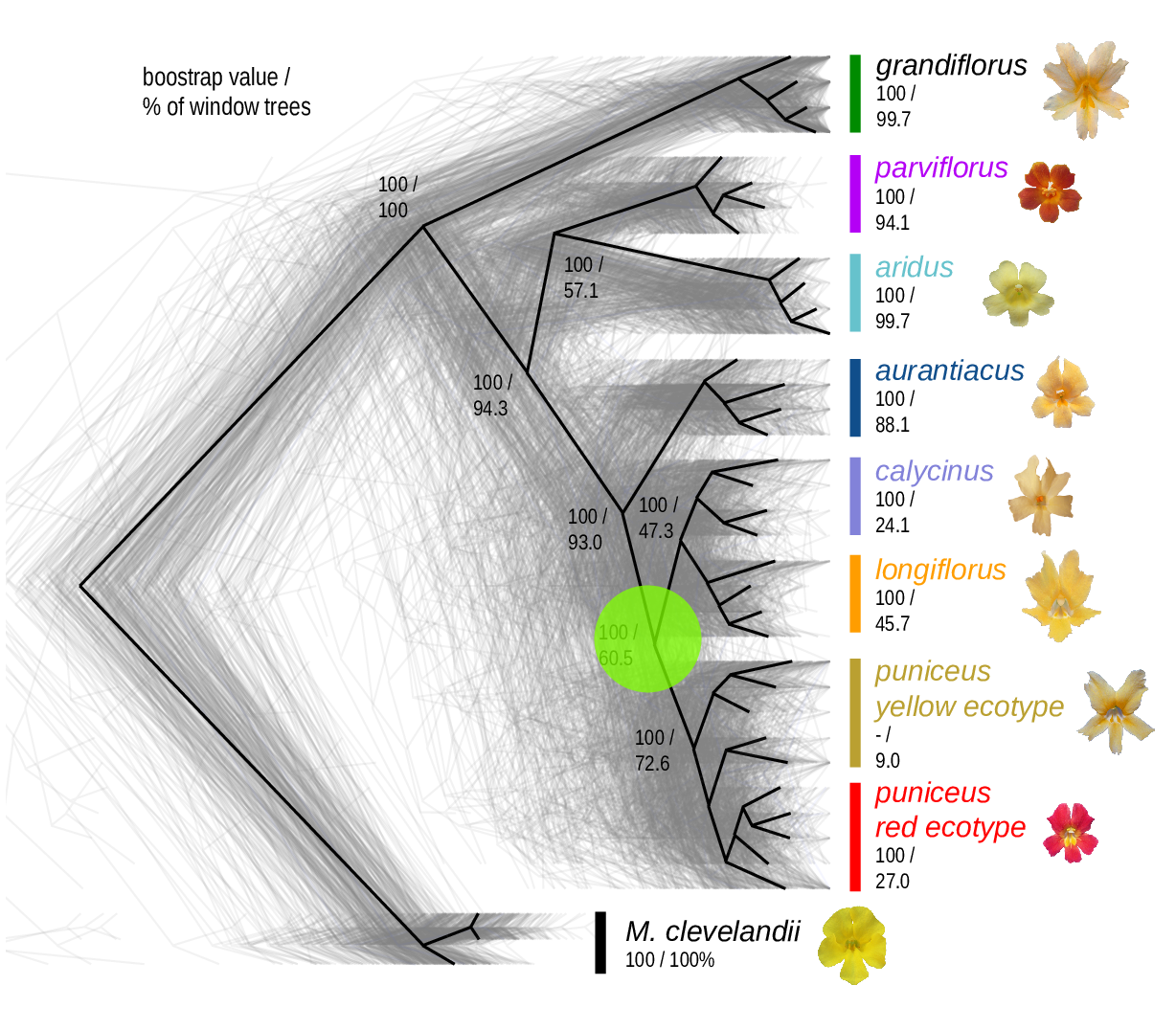
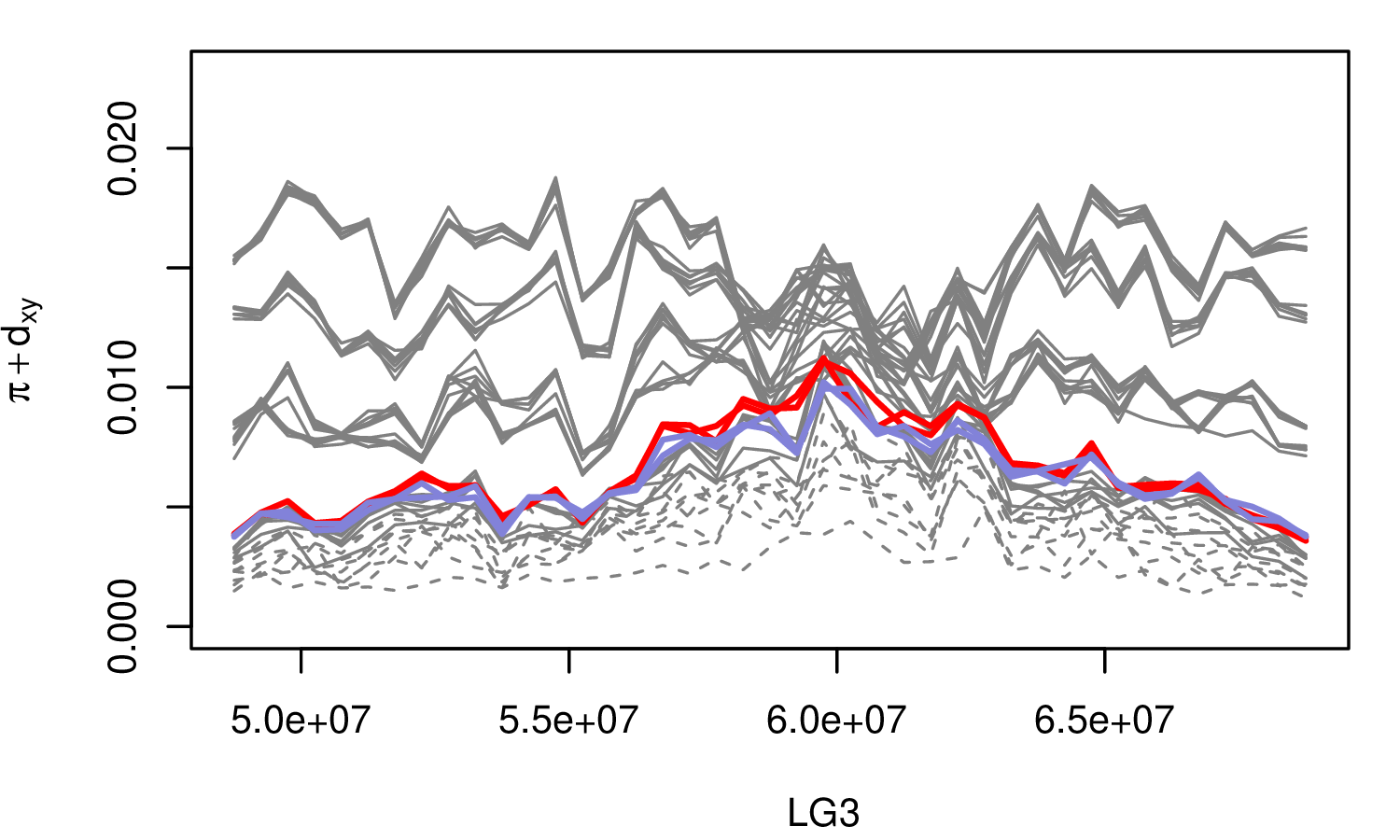
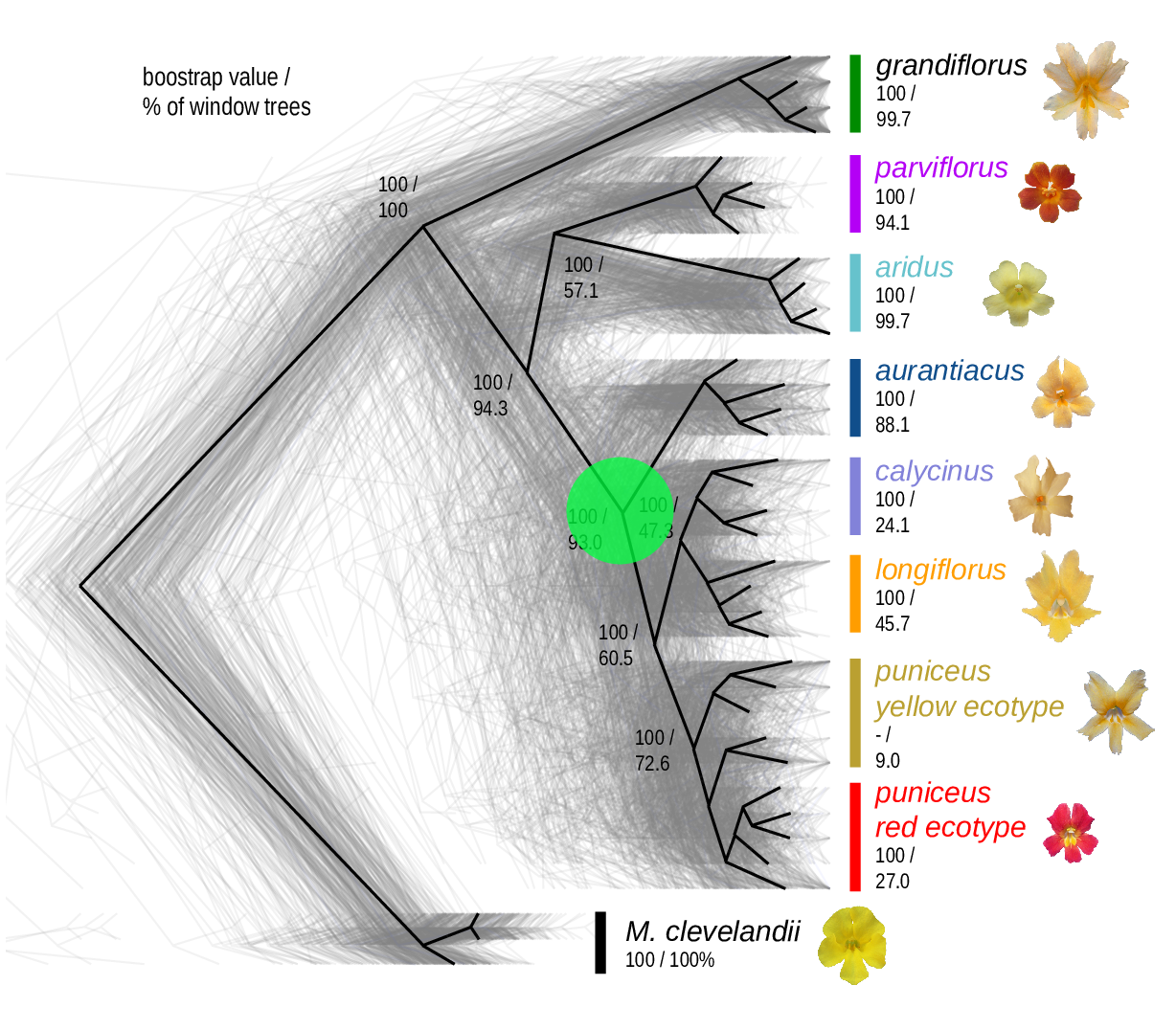
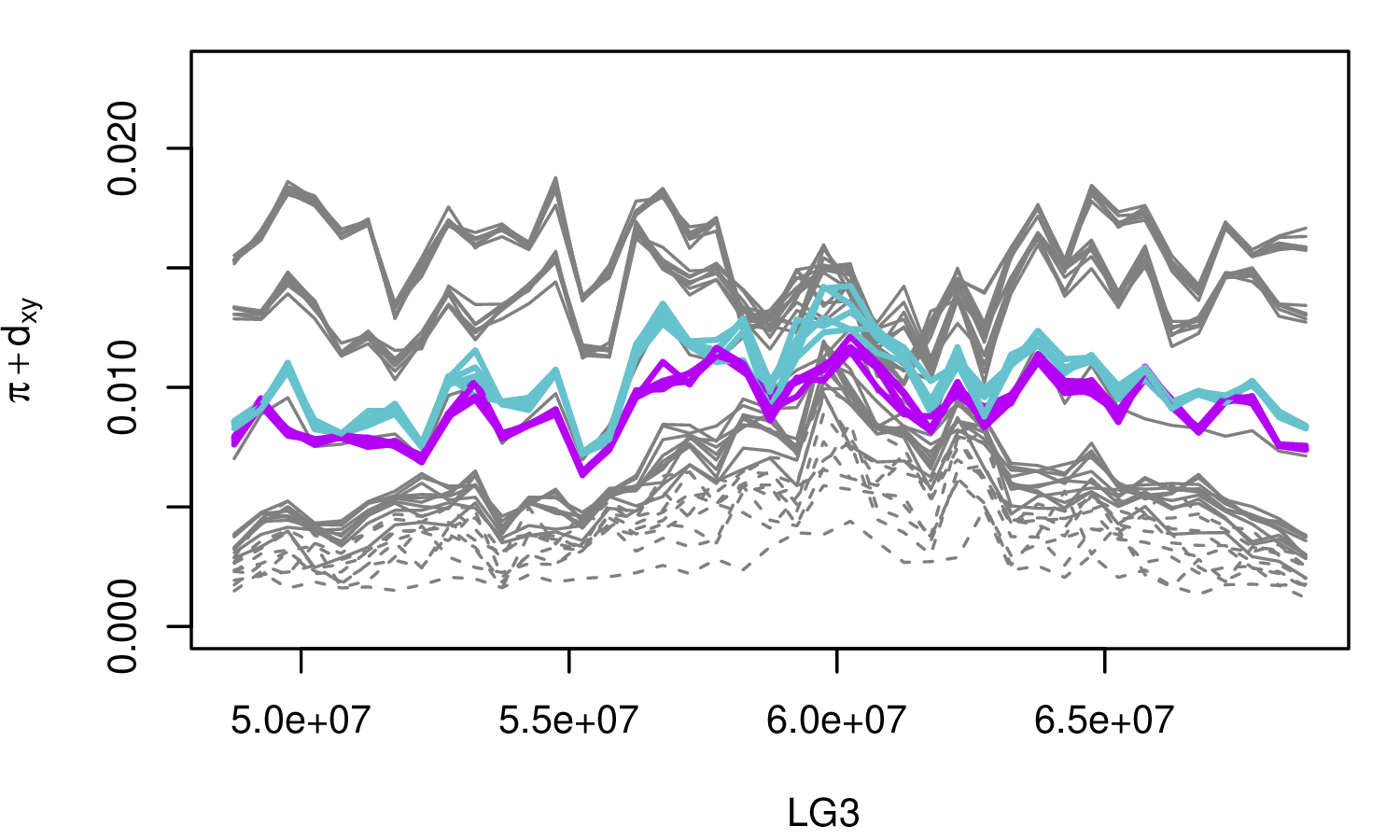
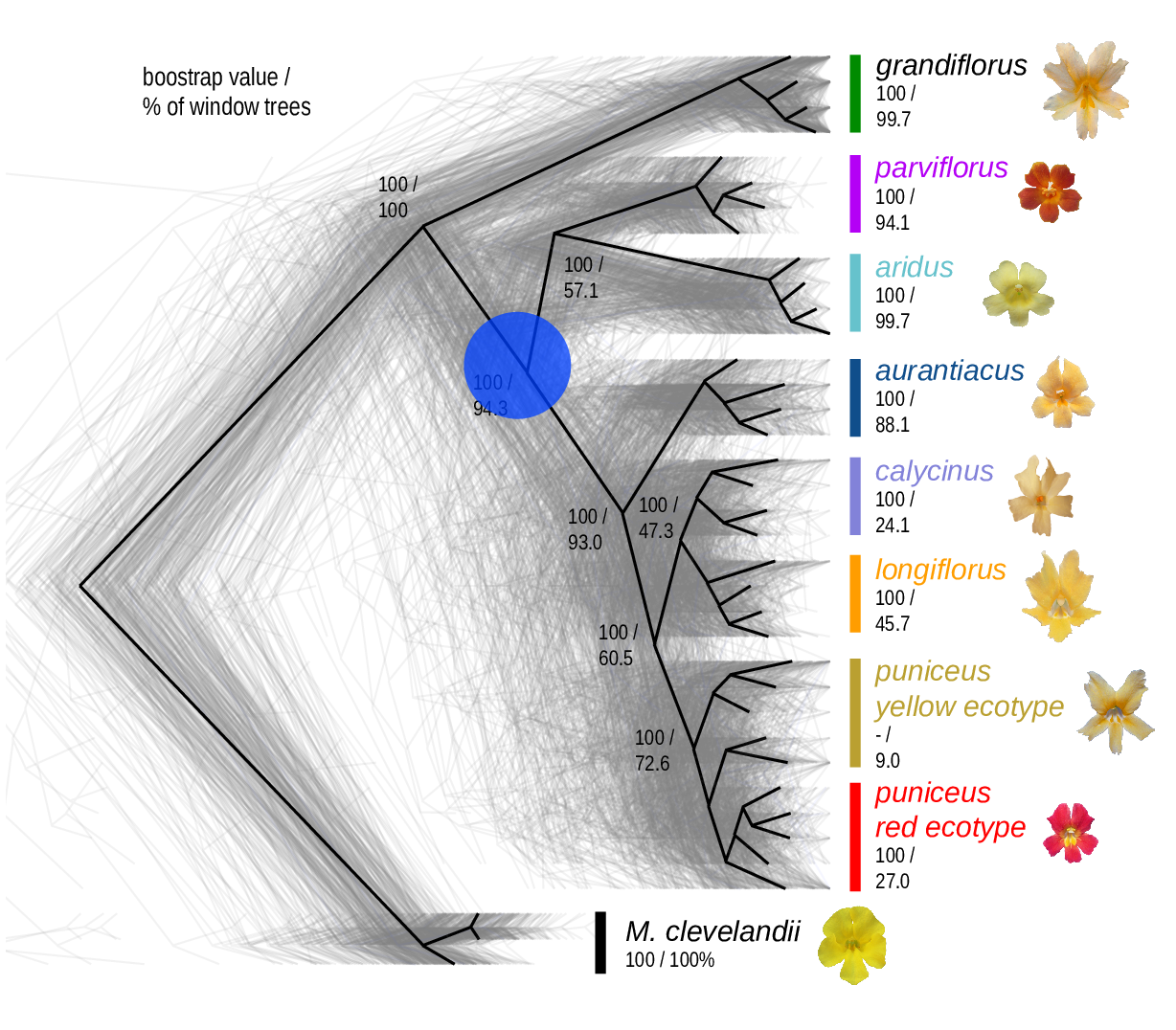
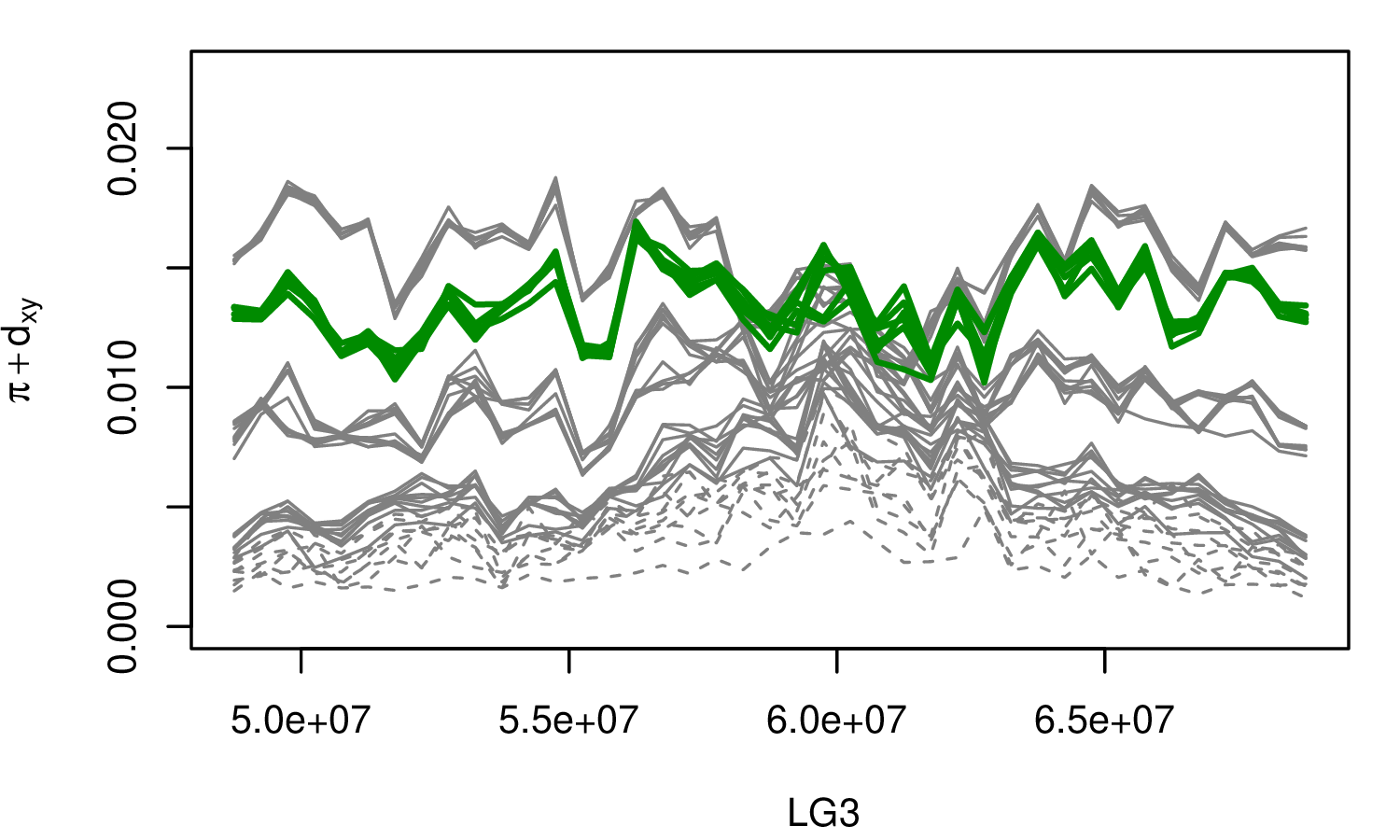
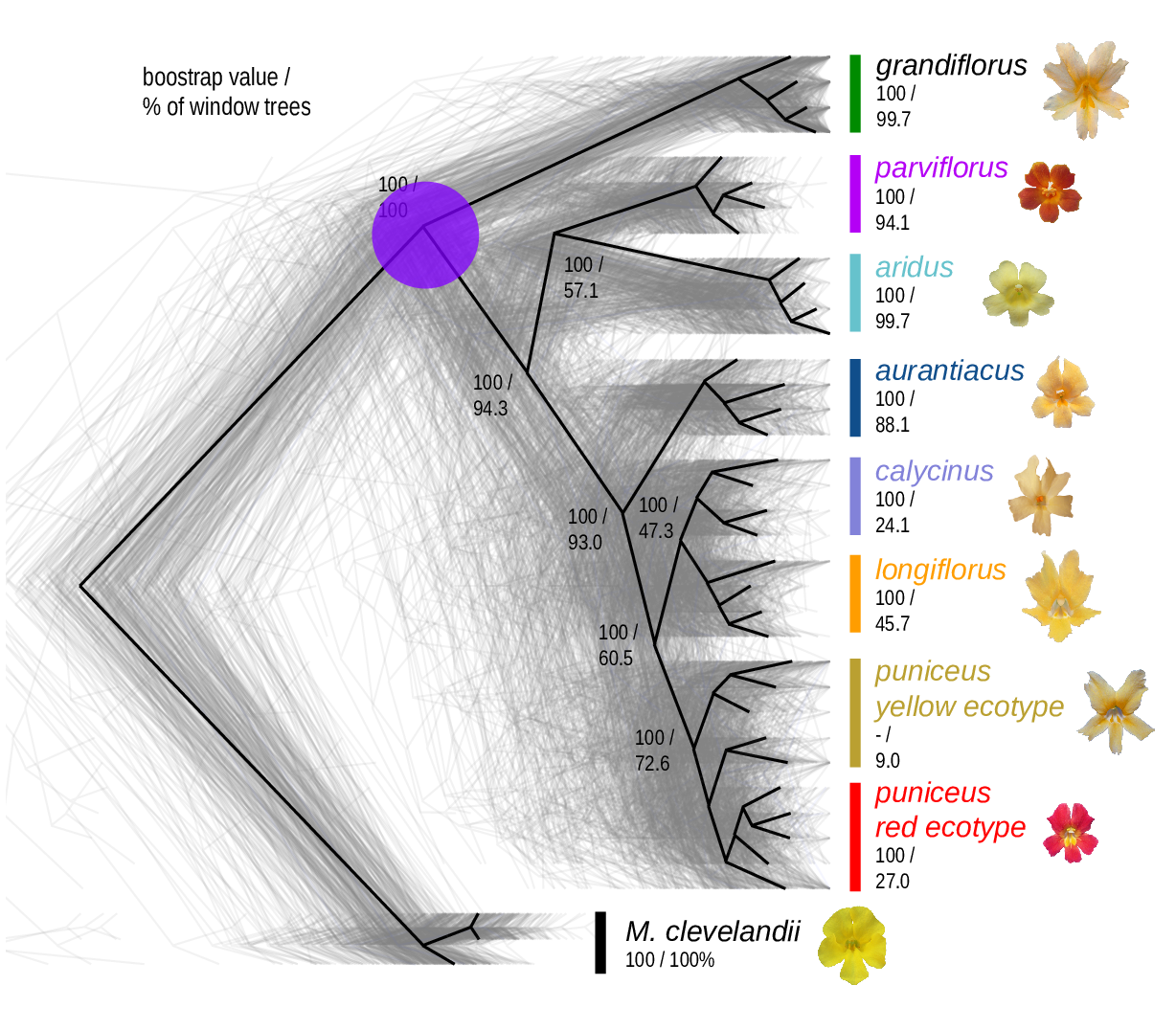
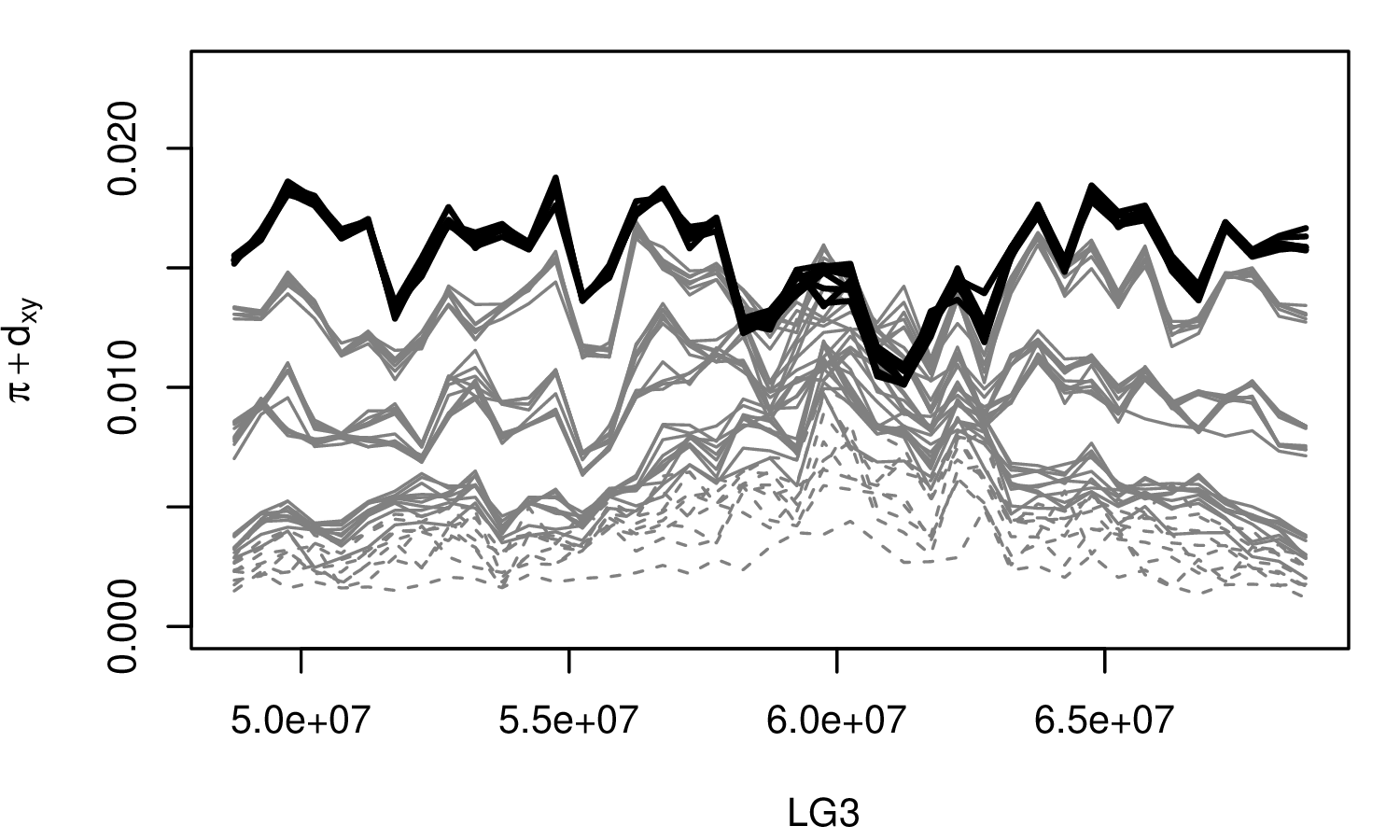
Some questions
Ok, then: selection.
But: what kind of selection?
- newly adaptive variants?
- purifying selection?
- local adaptation?
- selection for introgression?
To test theories and fit models, we need simulations with realistic
- population sizes,
- genomes,
- selective pressures,
- histories, and
- geography.
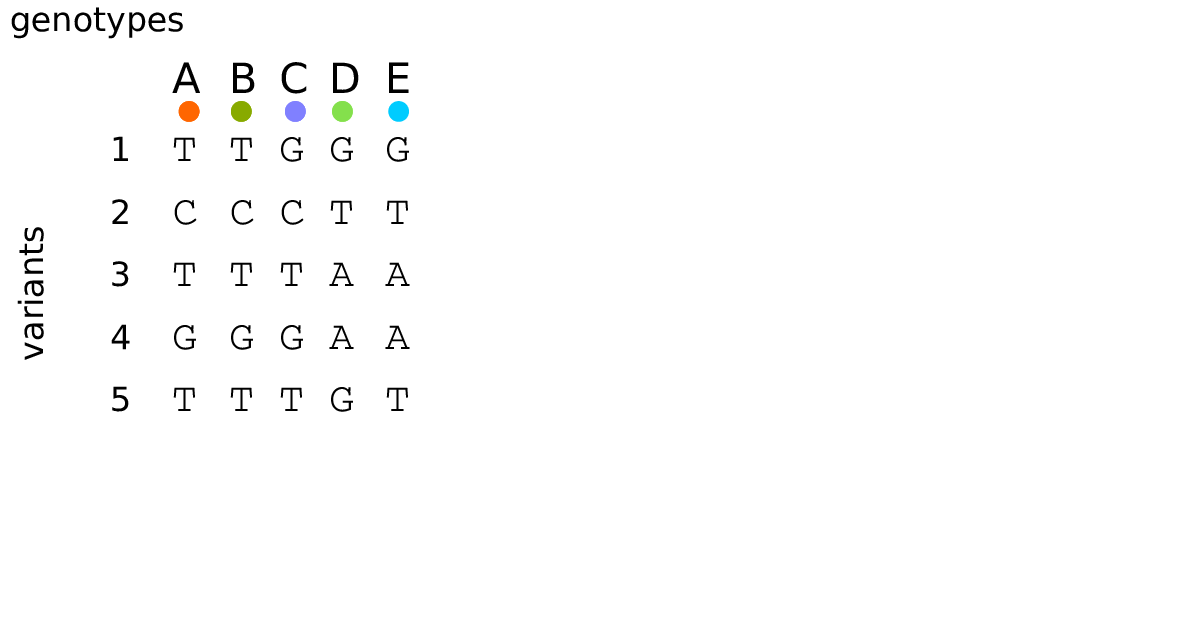
The tree sequence
History is a sequence of trees
For a set of sampled chromosomes, at each position along the genome there is a genealogical tree that says how they are related.
The tree sequence is a way to describe this, er, sequence of trees.
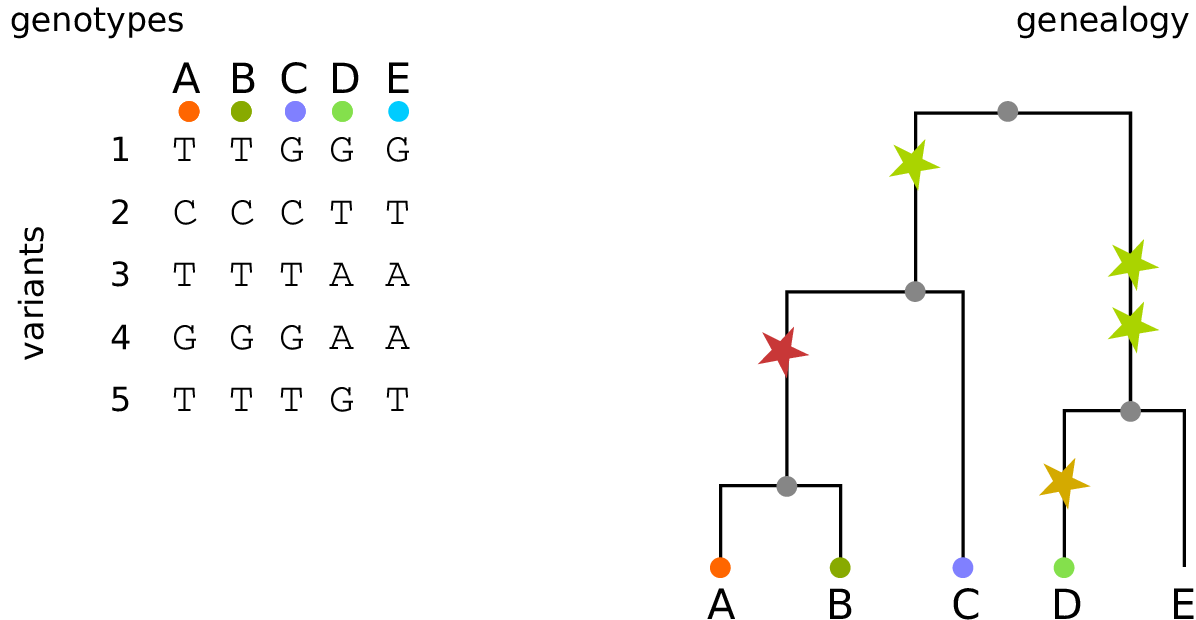
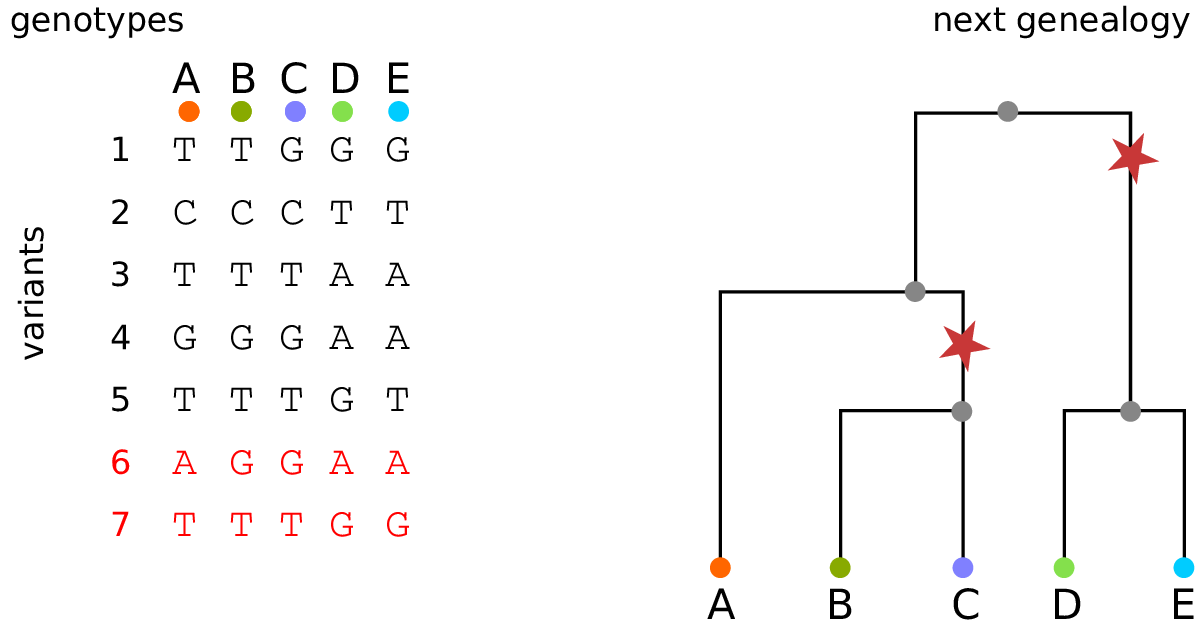
Kelleher, Etheridge, and McVean introduced the tree sequence data structure for a fast coalescent simulator, msprime.
stores sequence and genealogical data very efficiently
tree-based sequence storage closely related to haplotype-matching compression
tskit: python/C tools


jerome kelleher
File sizes
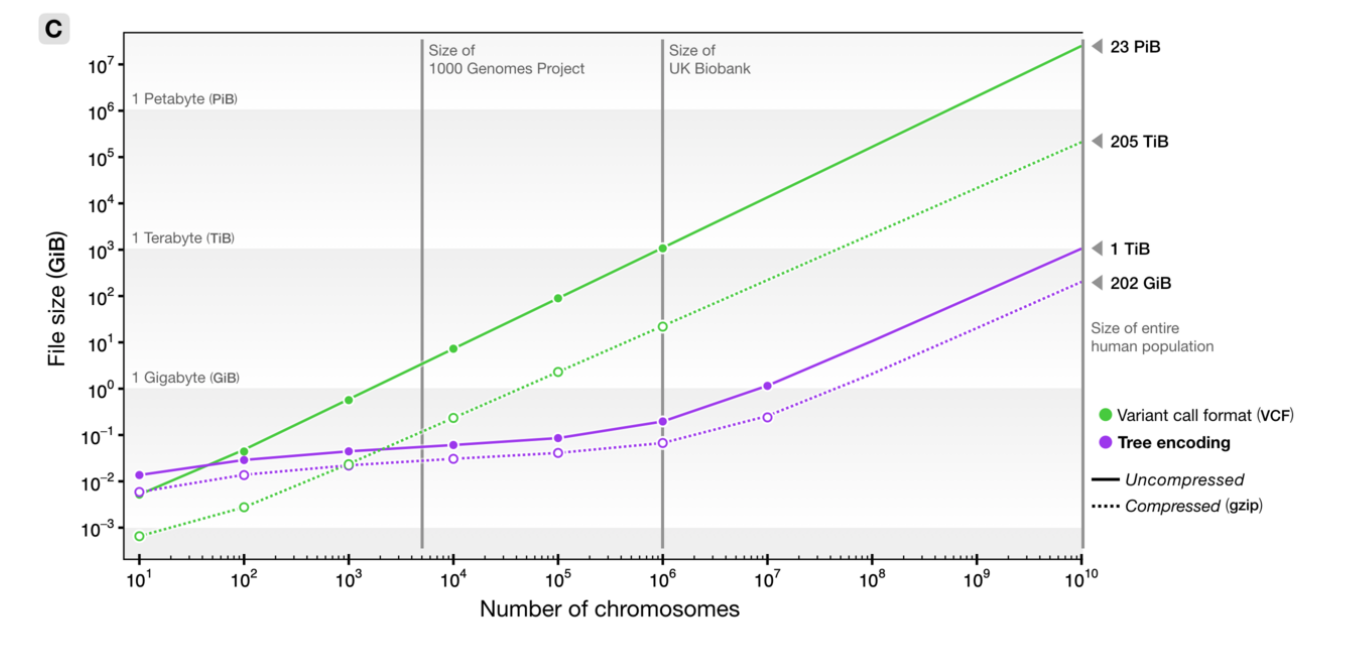
from Kelleher et al 2018, Inferring whole-genome histories in large population datasets, Nature Genetics
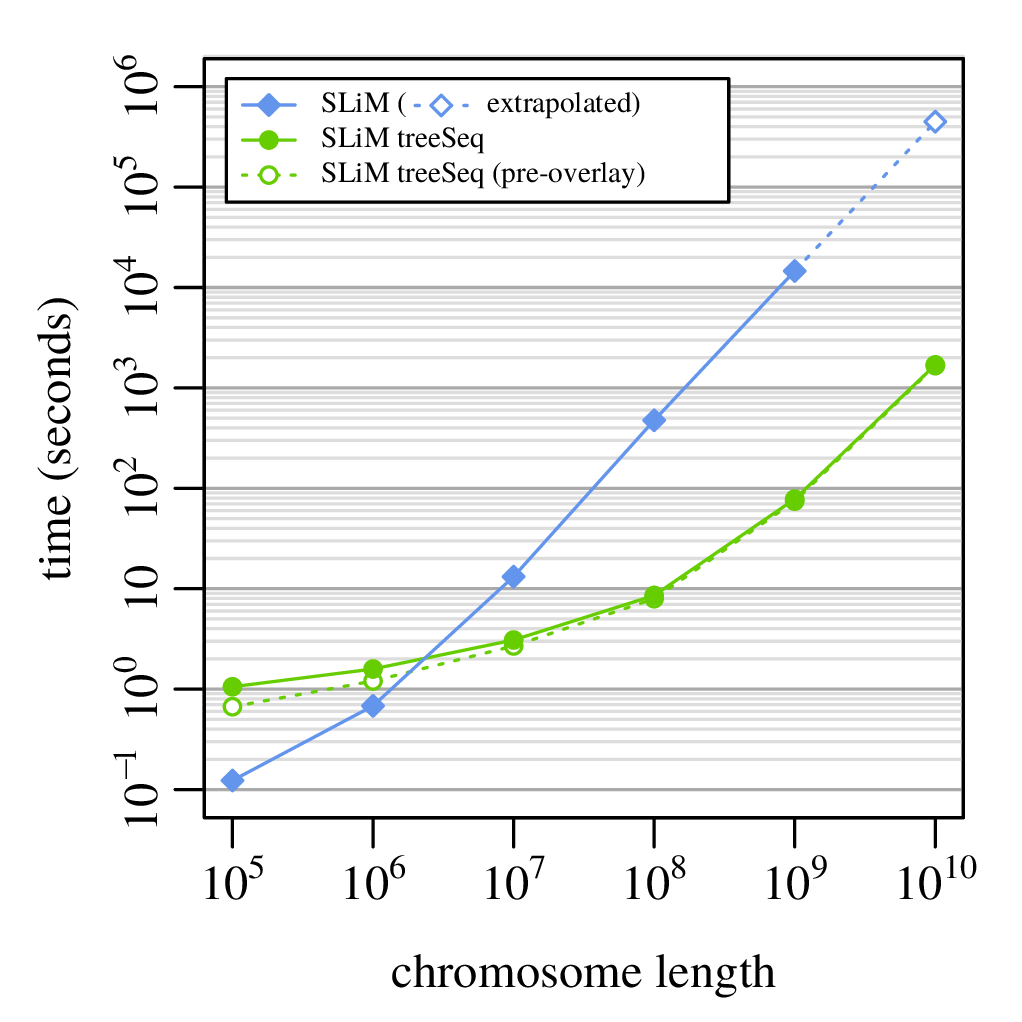
Computation run time
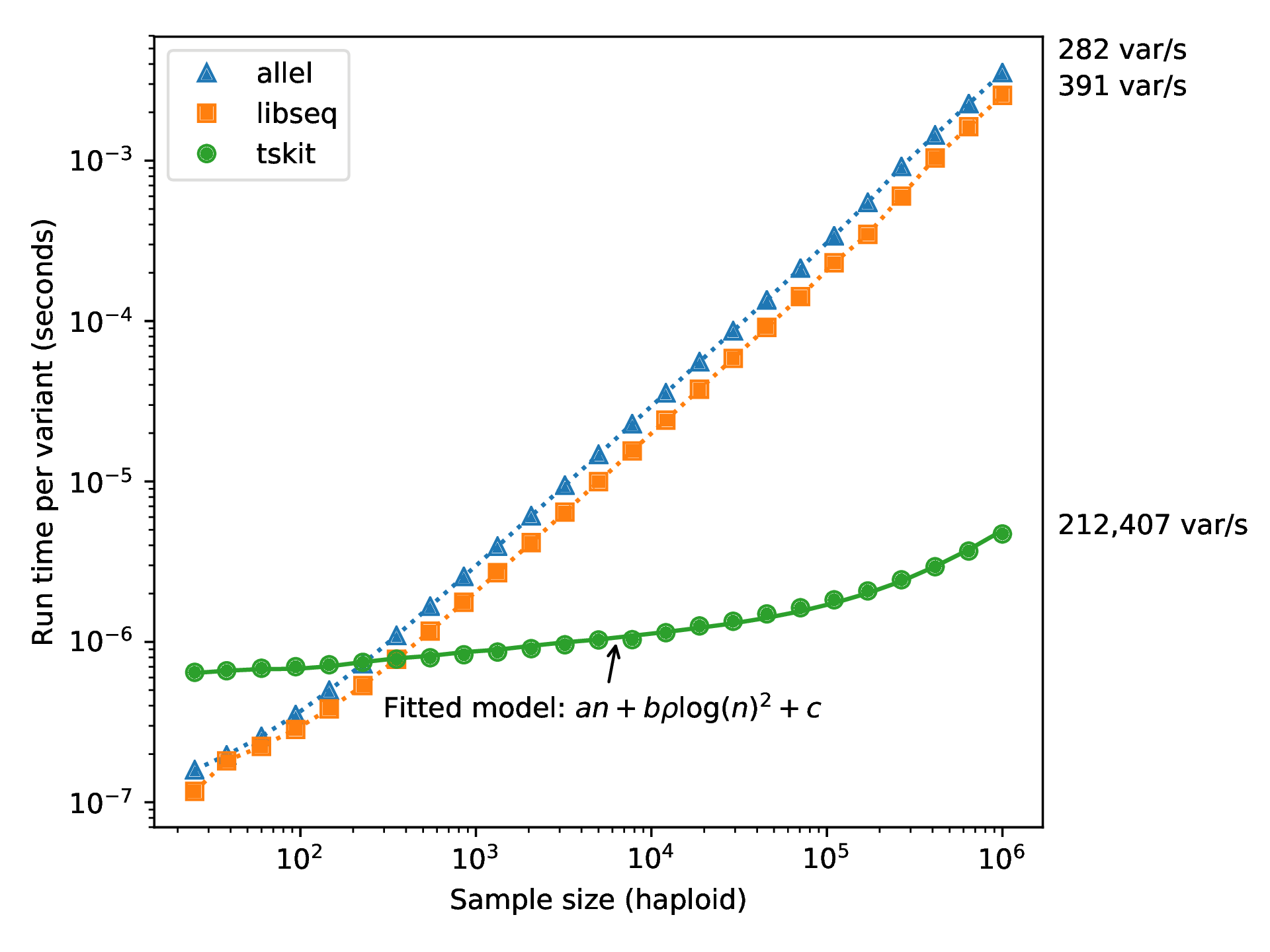
from Ralph, Thornton and Kelleher 2019, Efficiently summarizing relationships in large samples
Application to genomic simulations
The main idea
If we record the tree sequence that relates everyone to everyone else,
after the simulation is over we can put neutral mutations down on the trees.
Since neutral mutations don’t affect demography,
this is equivalent to having kept track of them throughout.
From Kelleher, Thornton, Ashander, and Ralph 2018, Efficient pedigree recording for fast population genetics simulation.
and Haller, Galloway, Kelleher, Messer, and Ralph 2018, Tree‐sequence recording in SLiM opens new horizons for forward‐time simulation of whole genomes



This means recording the entire genetic history of everyone in the population, ever.
It is not clear this is a good idea.
But, with a few tricks…
A 100x speedup!
What else can you do with tree sequences?
- recorded pedigree and migration history
- true ancestry assignment
- recapitation: fast, post-hoc initialization with coalescent simulation
- fast, convenient computation
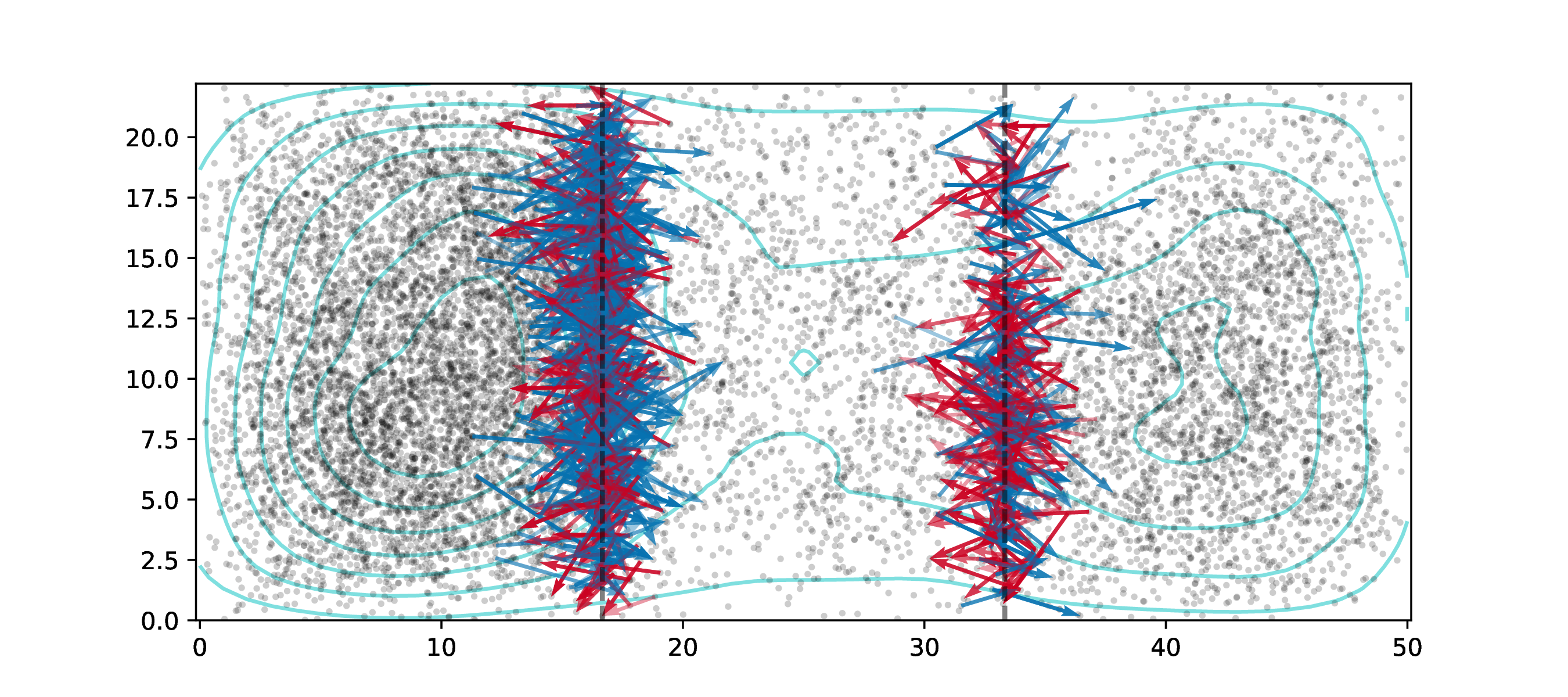
For example:
- genome as human chr7 (\(1.54 \times 10^8\)bp)
- \(\approx\) 10,000 diploids
- 500,000 overlapping generations
- continuous, square habitat
- selected mutations at rate \(10^{-10}\)
- neutral mutations added afterwards
Runtime: 8 hours
Back to Mimulus
The data


Simulations
\(N=10,000\) diploids
burn-in for \(10N\) generations
population split, with either:
- neutral
- background selection
- selection against introgressed alleles
- positive selection
- local adaptation

Murillo Rodrigues
From Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers, Stankowski, Chase, Fuiten, Rodrigues, Ralph, and Streisfeld; PLoS Bio 2019.
Conclusions:
neutralbackground selectionselection against introgressed alleles- positive selection
- local adaptation
Wrap-up
Bigger picture
How strongly does selection enhance or constrain genetic variation?
How much genetic variation is locally adaptive?
How will populations respond to changes in the future?
Other uses for population simulation
train inference methods
predict management outcomes
do power analyses
develop intuition


The Co-Lab:
- Andy Kern
- Matt Lukac
- Murillo Rodrigues
- Jared Galloway
- CJ Battey
Funding:
- NIH NIGMS
- NSF DBI
- Sloan foundation
- UO Data Science
Other collaborators:
- Jerome Kelleher
- Ben Haller
- Ben Jeffery
- Georgia Tsambos
- Jaime Ashander
- Gideon Bradburd
- Madeline Chase
- Bill Cresko
- Alison Etheridge
- Evan McCartney-Melstad
- Brad Shaffer
- Sean Stankowski
- Matt Streisfeld
- Anastasia Teterina