Landscapes in population genetics:
ecology, evolution, and simulation
Peter Ralph
University of Oregon // October 2019
Outline
Outline of the talk
- Big picture
- Tools
- Applications
Students/postdocs/researchers:
- Matt Lukac
- Murillo Rodrigues
- Jared Galloway
- Jaime Ashander
- Josh Schiffman
- Erik Lundgren
- Han Li
- Jessica Crisci
Funding:
- NSF DBI
- Sloan foundation
- UO Data Science
Other collaborators:
- CJ Battey
- Gideon Bradburd
- Yaniv Brandvain
- Madeline Chase
- Graham Coop
- Bill Cresko
- Matt Dean
- Alison Etheridge
- Ben Haller
- Katja Kasimatis
- Jerome Kelleher
- Andy Kern
- Evan McCartney-Melstad
- Patrick Phillips
- Alisa Sedghifar
- Brad Shaffer
- Sean Stankowski
- Matt Streisfeld
- Anastasia Teterina
Adaptation, and genetic variation
Sickle-cell (HbS) allele frequencies
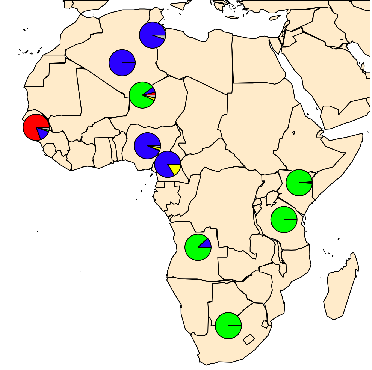
Human sickle-cell allele (HbS): (Currat et al 2002)
- Single base substitution
- provides protection against malaria (but deleterious in homozygotes)
G6PD deficiency allele frequencies
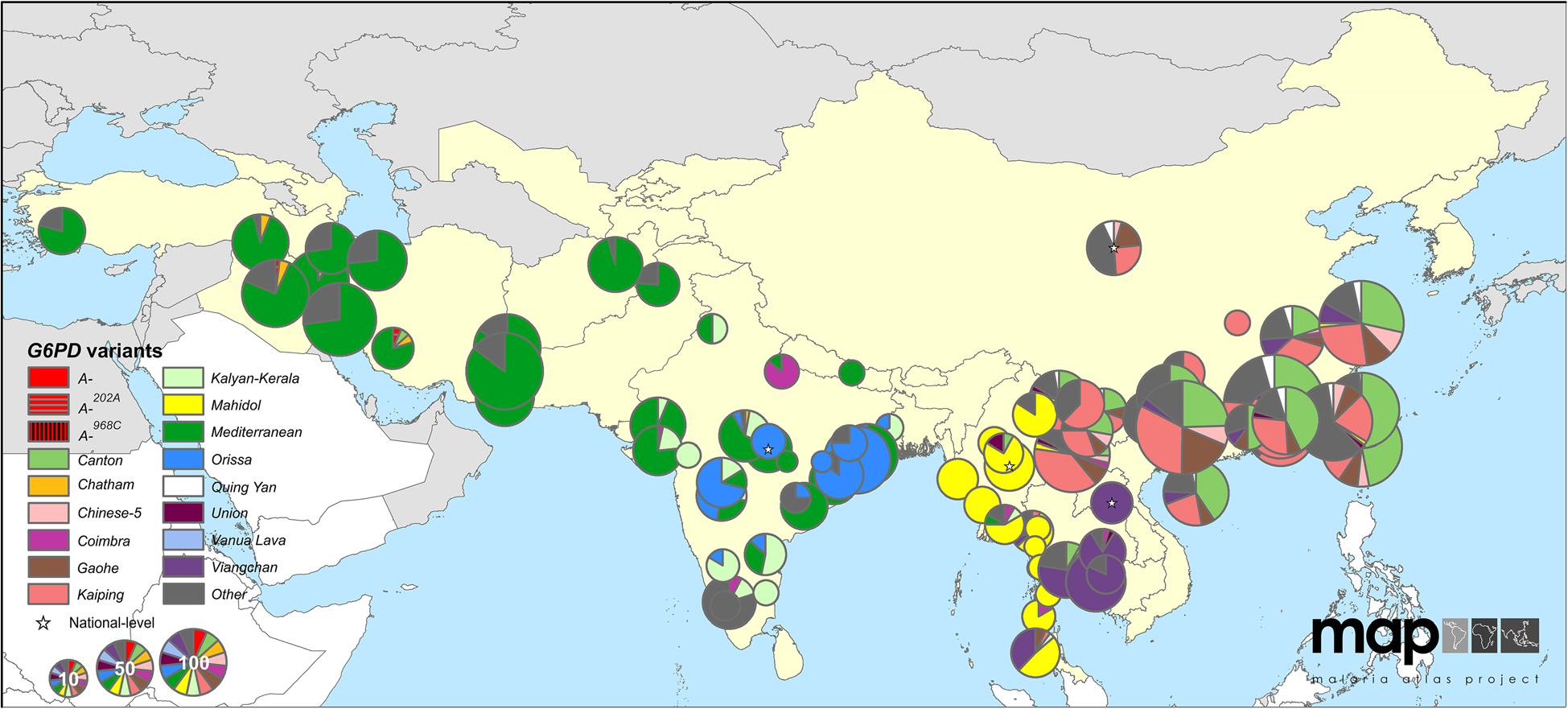
Human G6PD variants: Howes et al 2013)
- over 130 G6PD deficiency alleles; 34 variants at high frequency
- provide protection against malaria but increases risk of anemia
- Estimated ages 40-400 generations

- Dark-pigmented mammals and reptiles on volcanic outcrops in the Southwest. (Dice, Benson 1936)
- ‘Dark’ allele beneficial on outcrops, deleterious elsewhere.
- MC1R: basis is shared between species but not between populations (Nachman, Hoekstra)
How generalizable is this?
Genomic landscapes
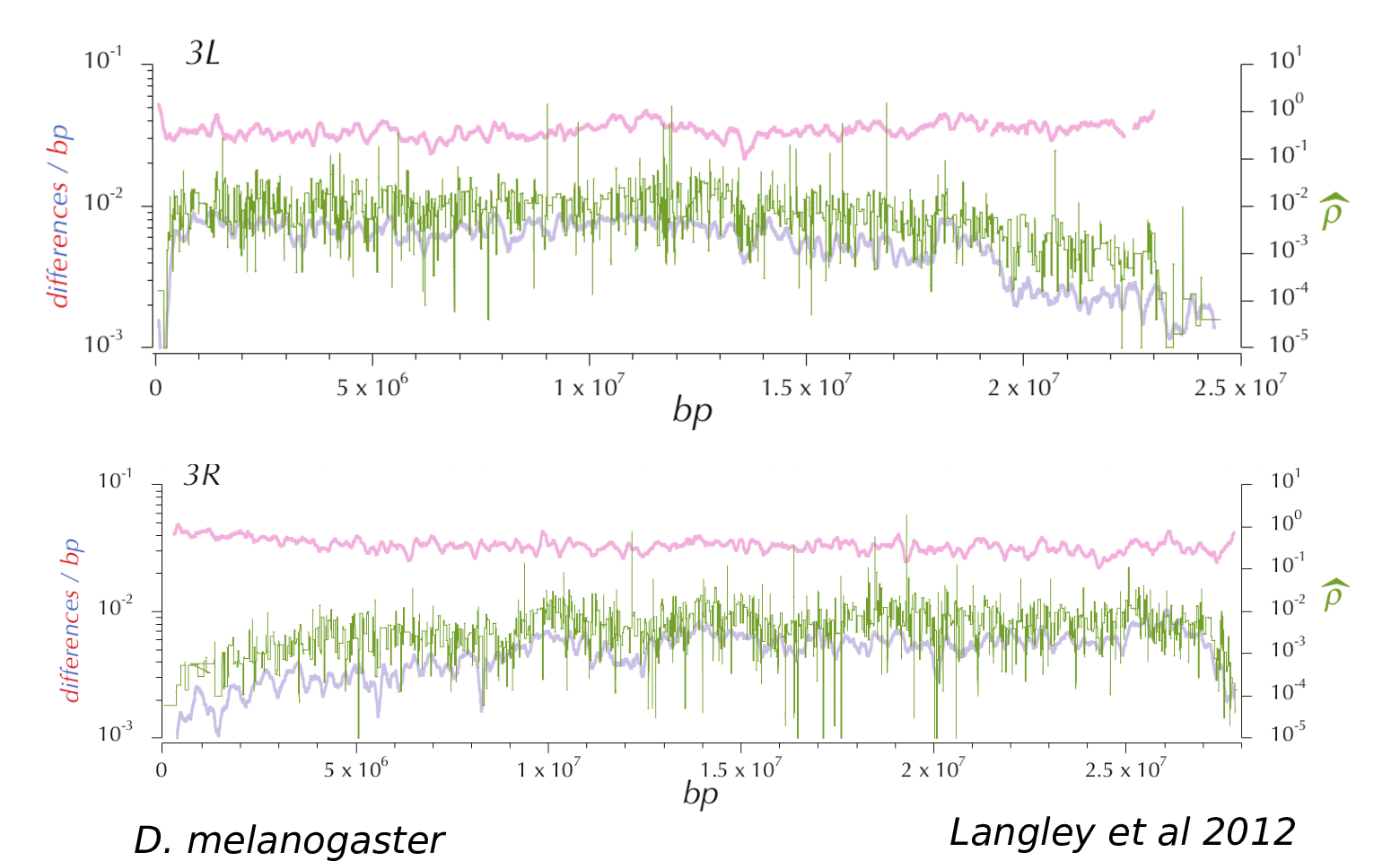
Diversity correlates with recombination rate
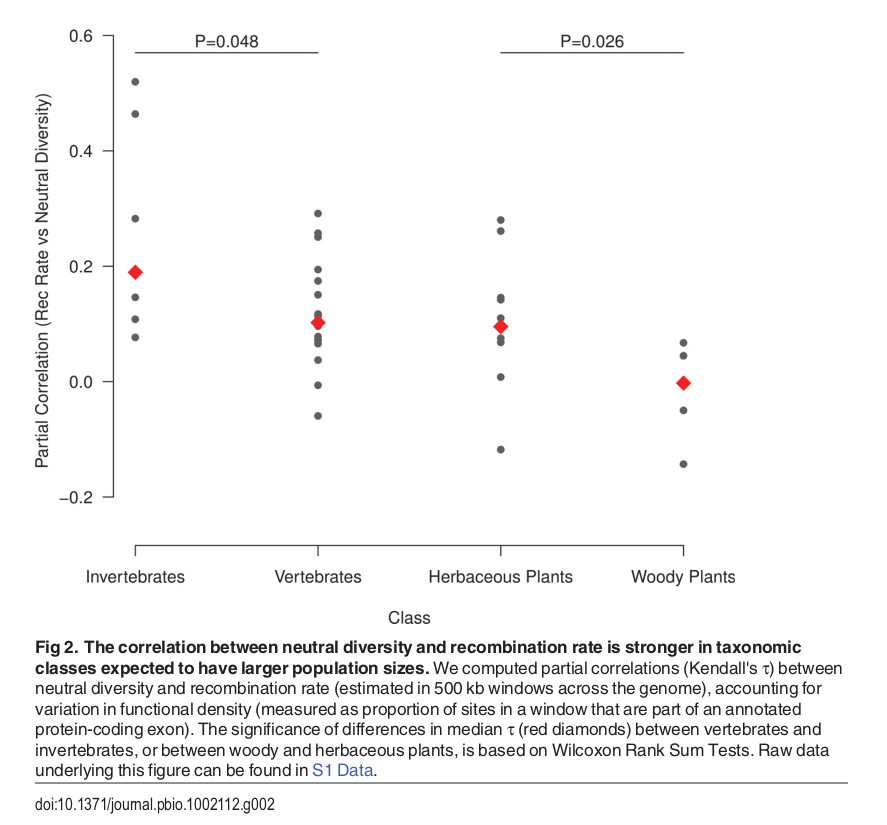
Hudson 1994; Cutter & Payseur 2013; Corbett-Detig et al 2015
- linked selection
The indirect effects of selection on genomic locations that are linked to the sites under selection by a lack of recombination.
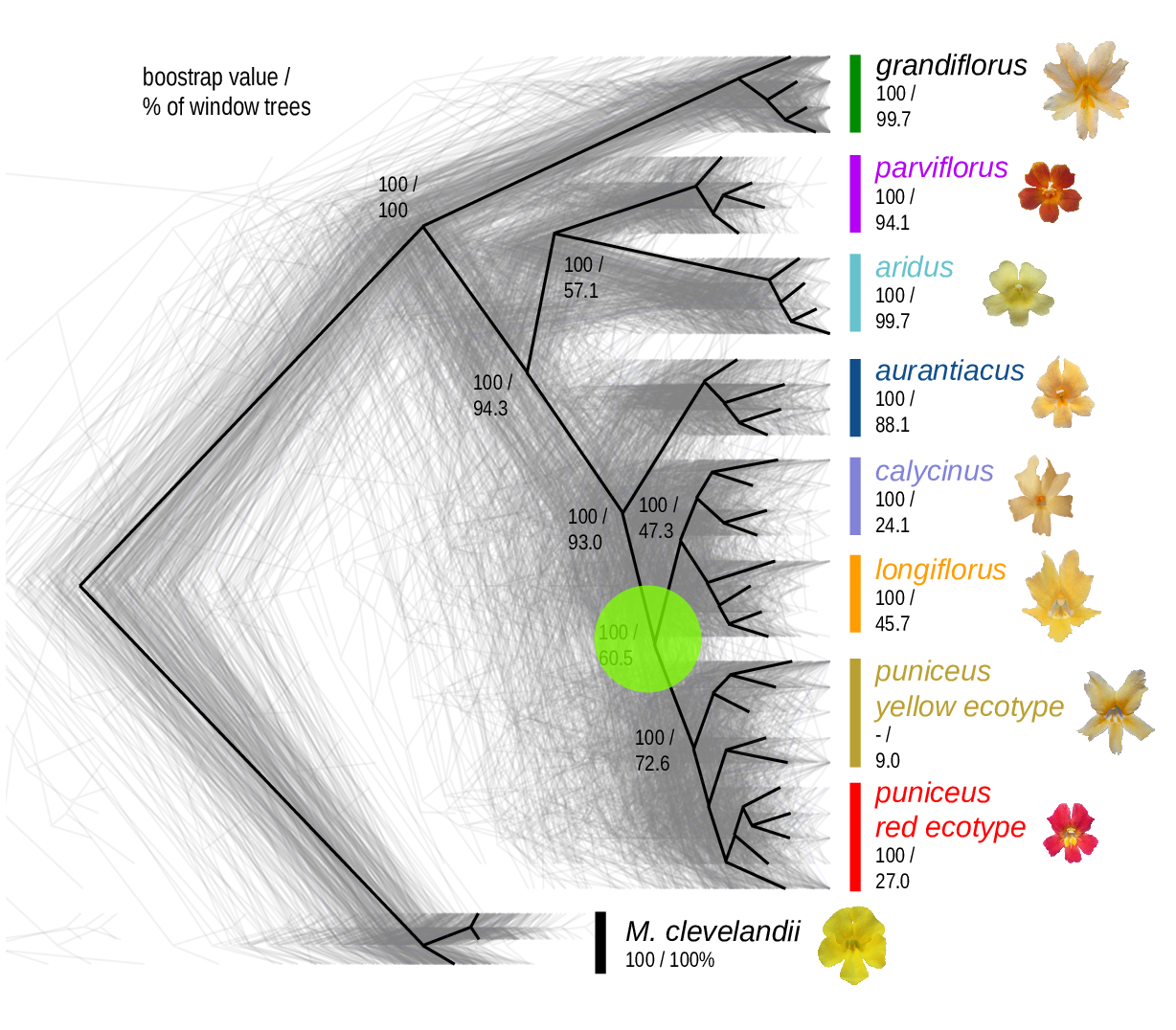
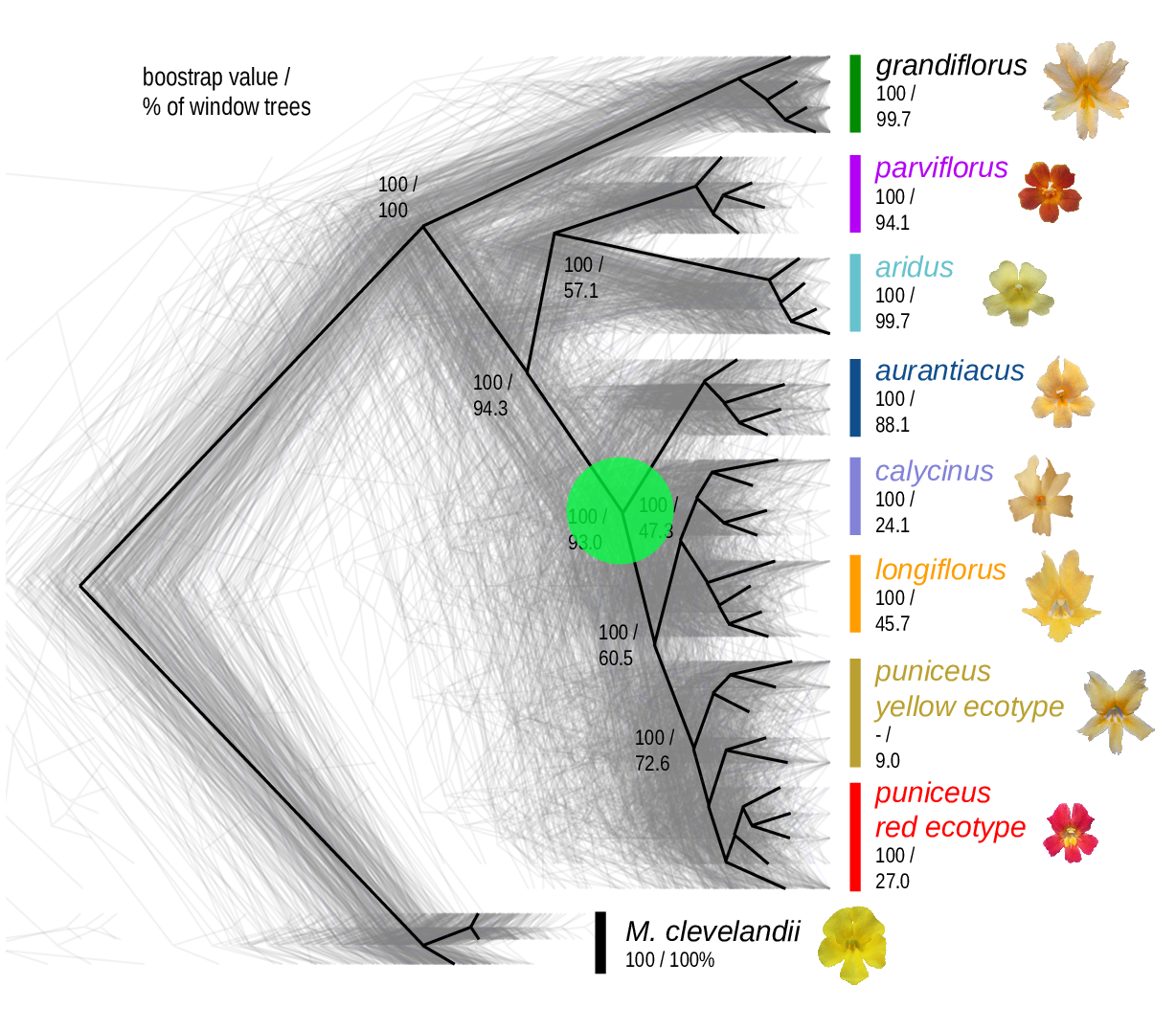
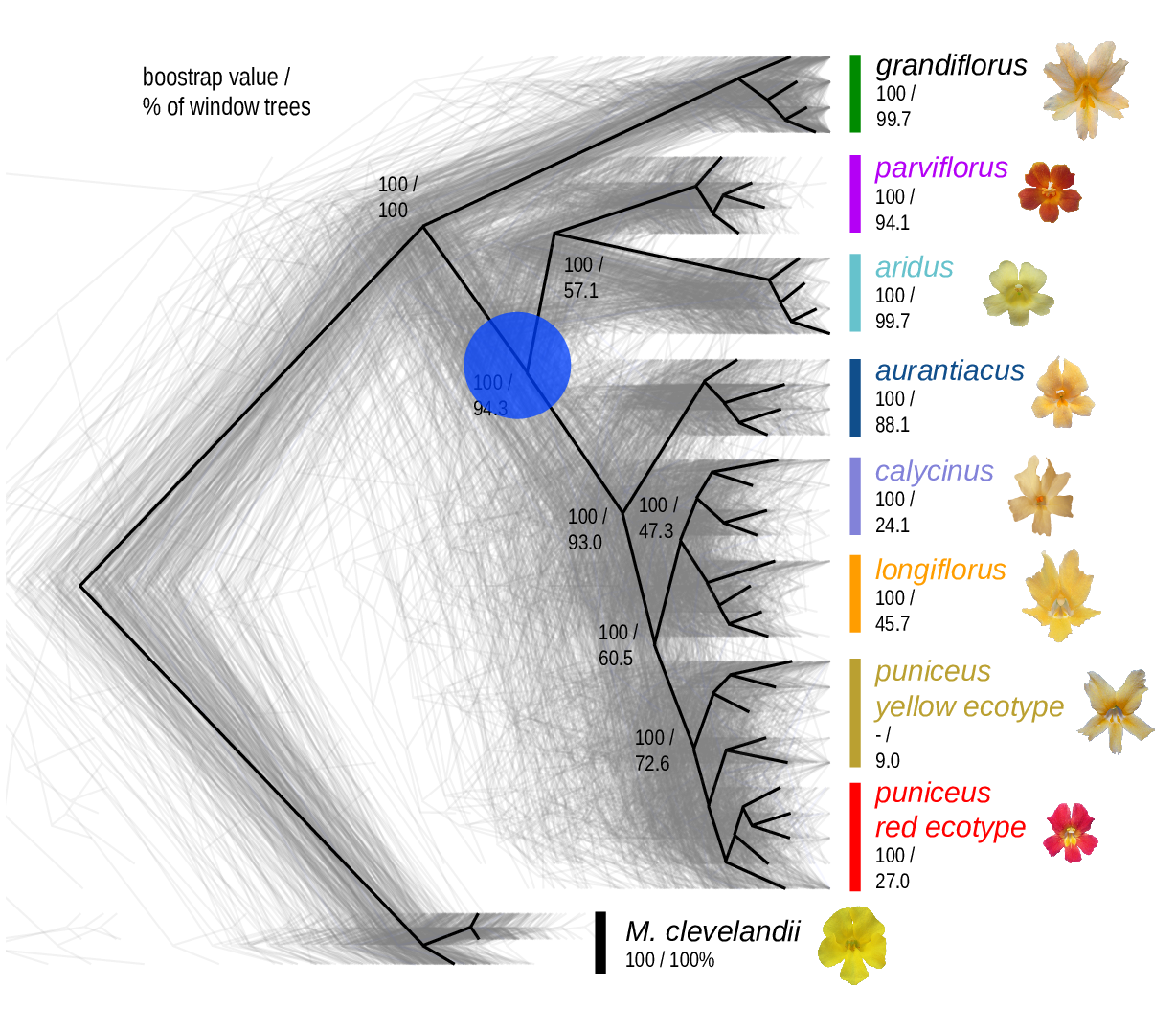
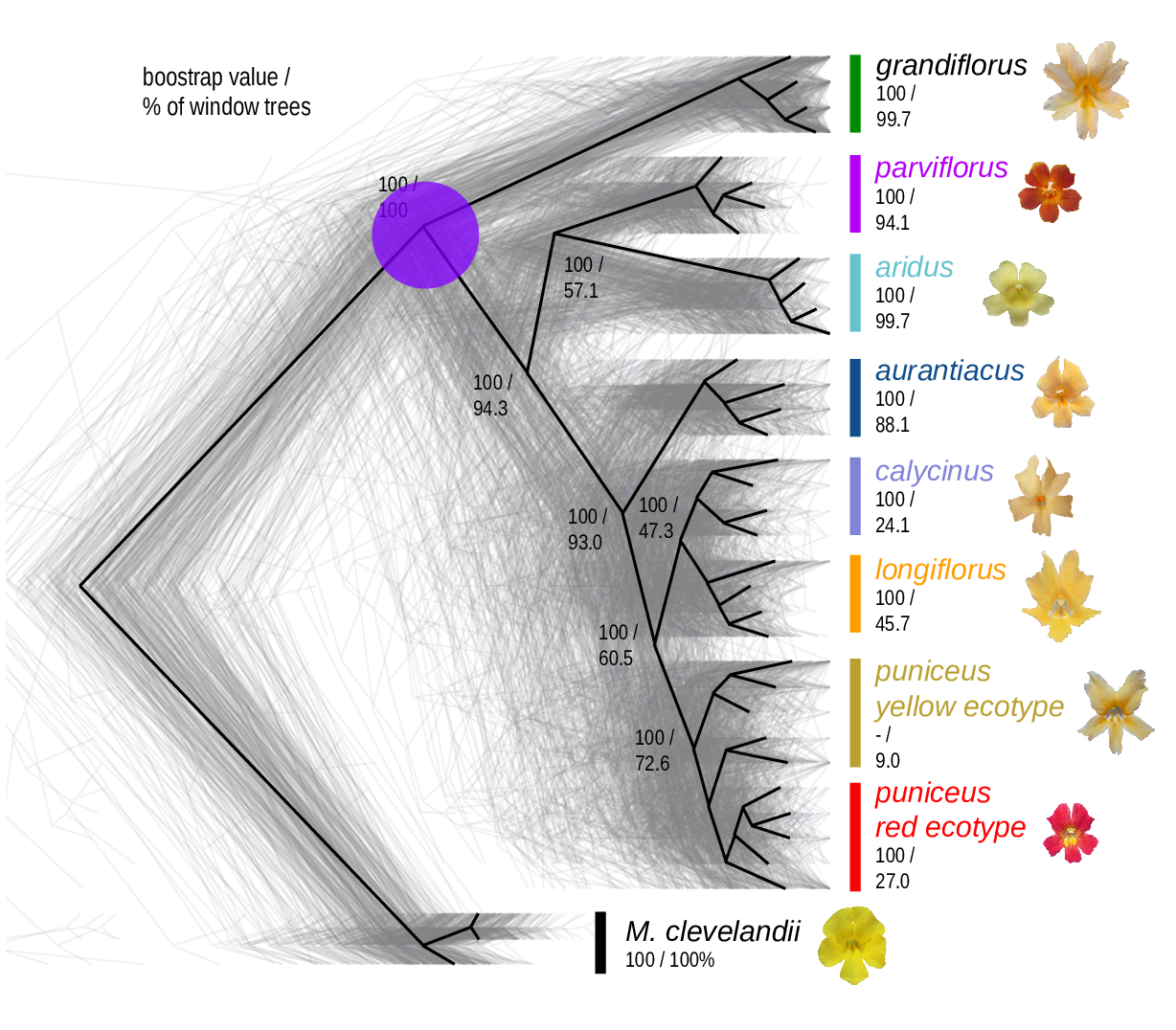
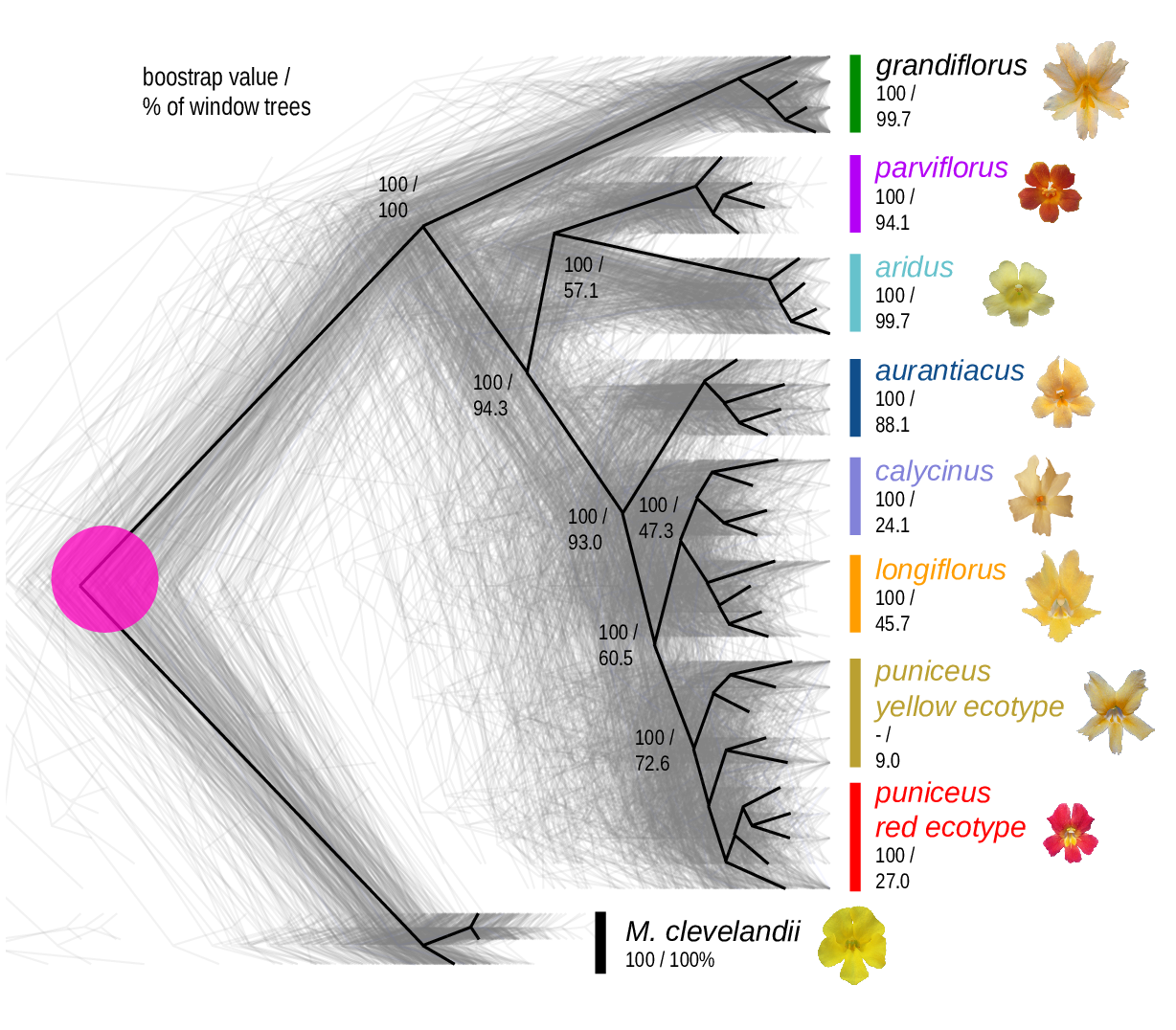
The Mimulus aurantiacus species complex
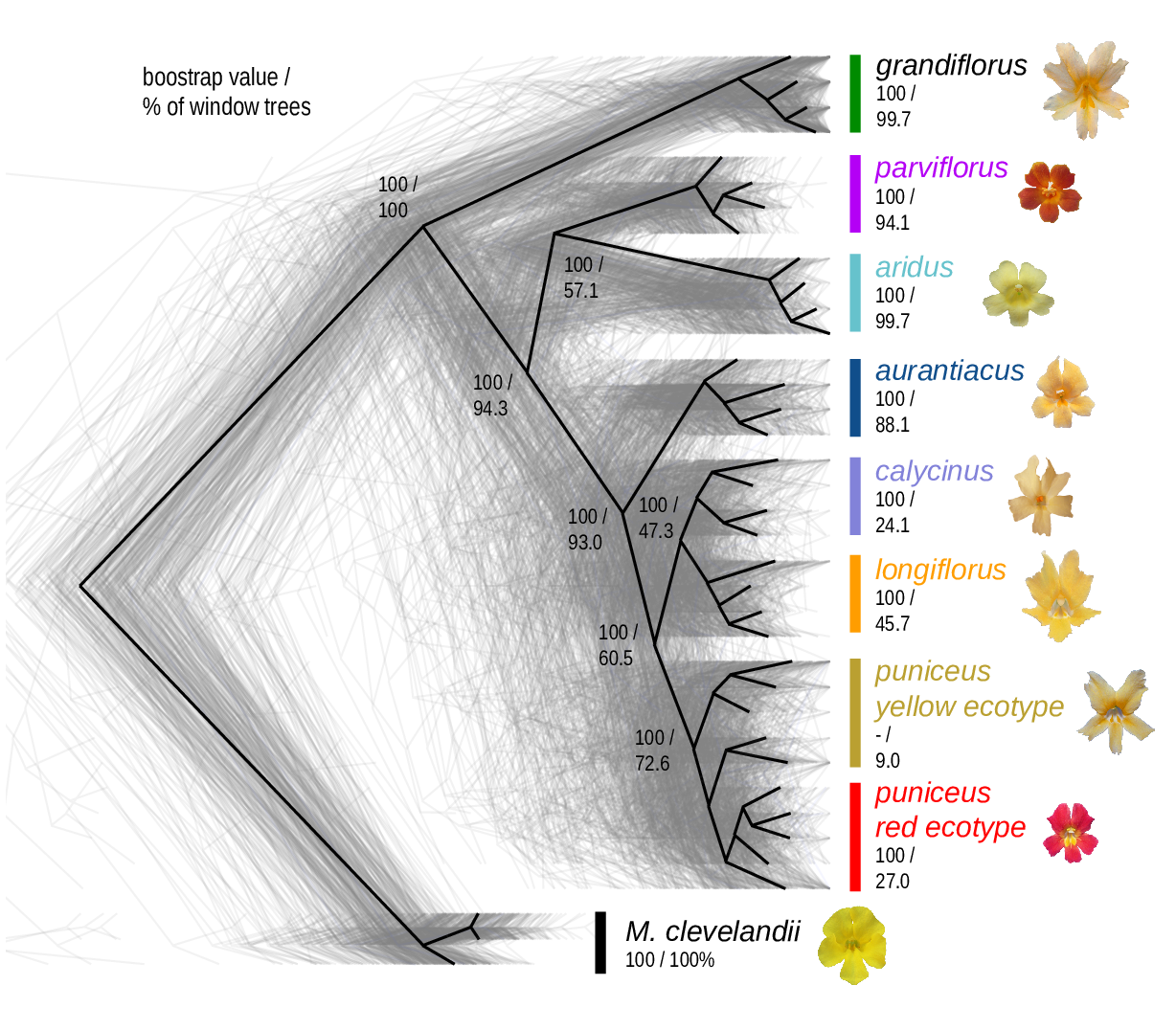
From Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers, Sean Stankowski, Madeline A. Chase, Allison M. Fuiten, Murillo F. Rodrigues, Peter L. Ralph, and Matthew A. Streisfeld; PLoS Bio 2019.



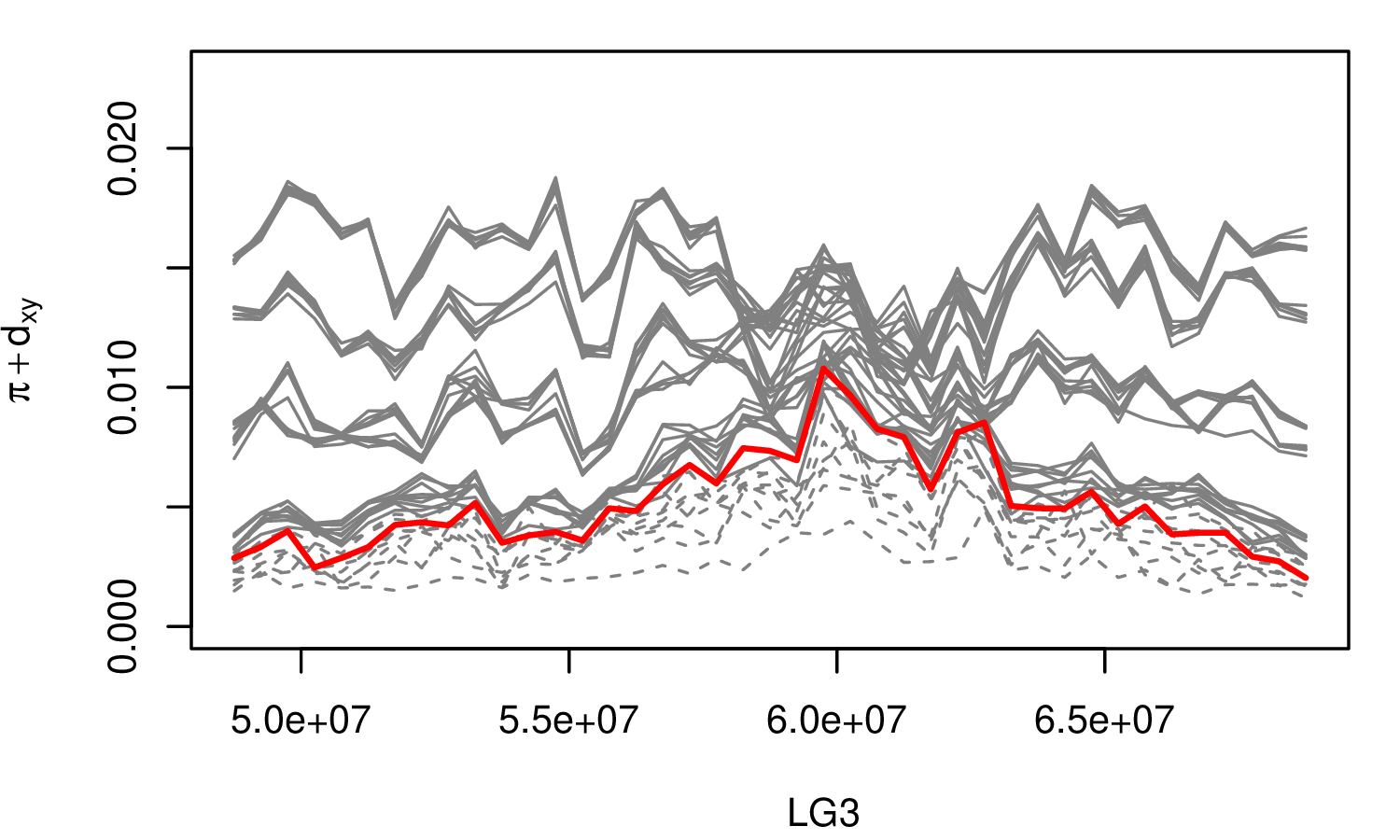
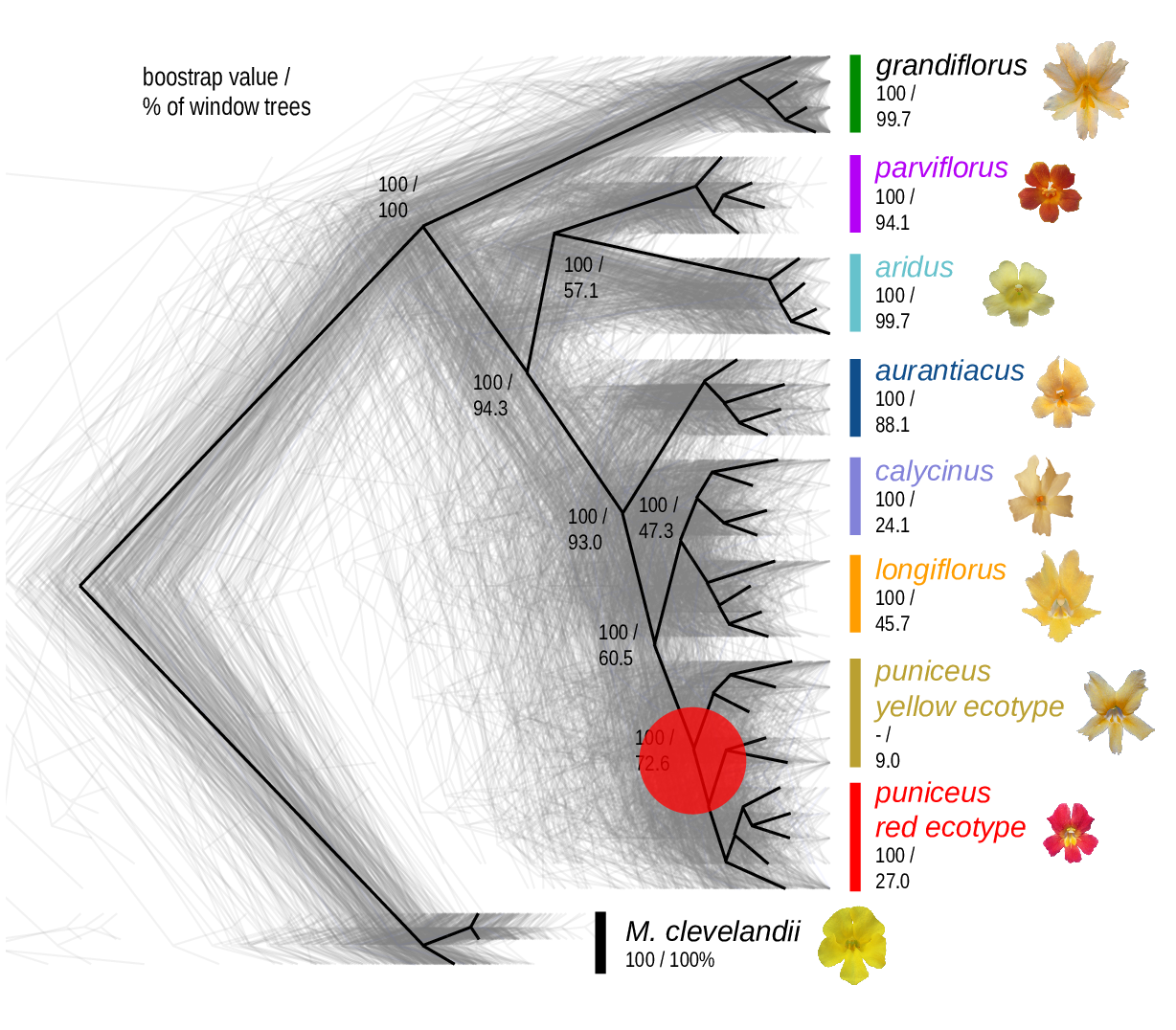
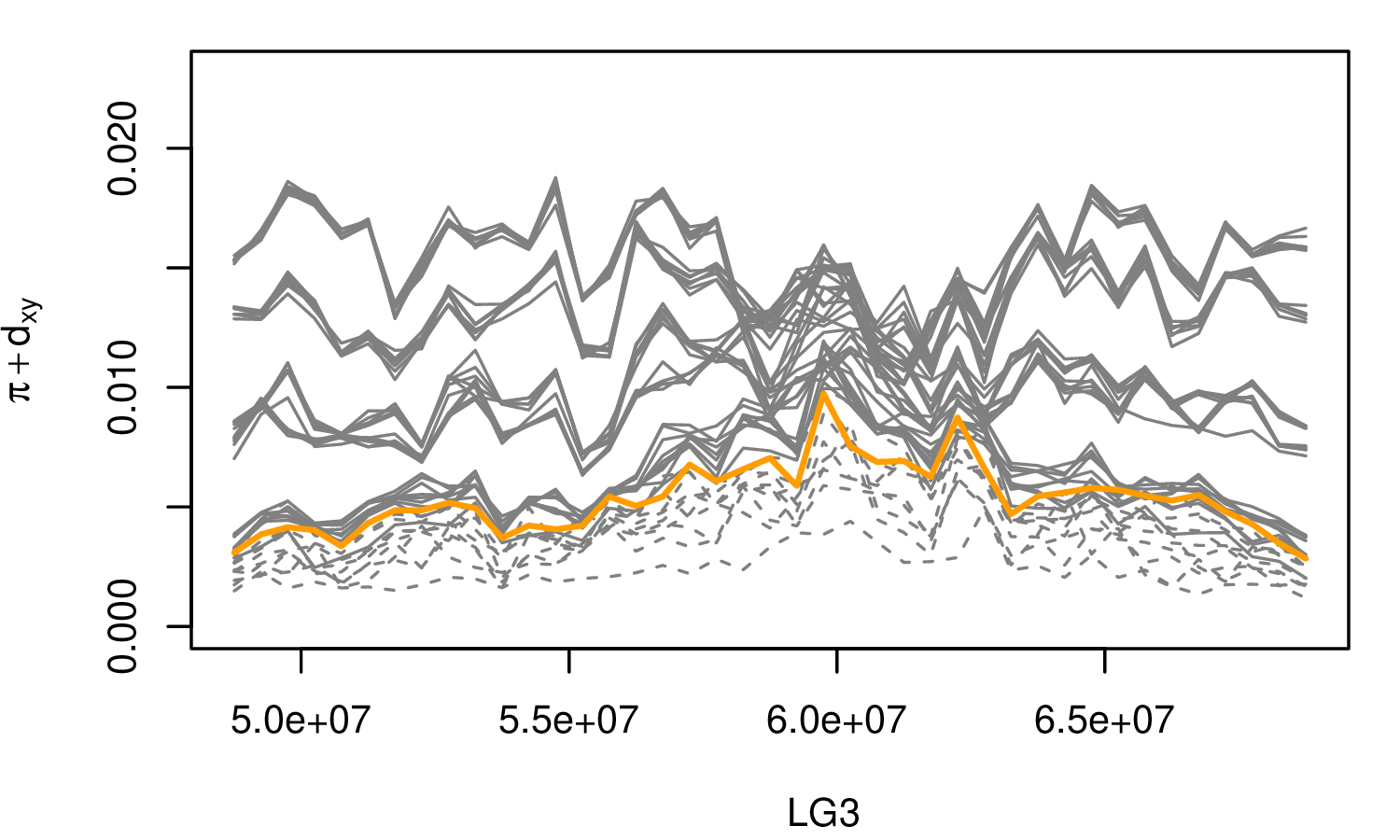
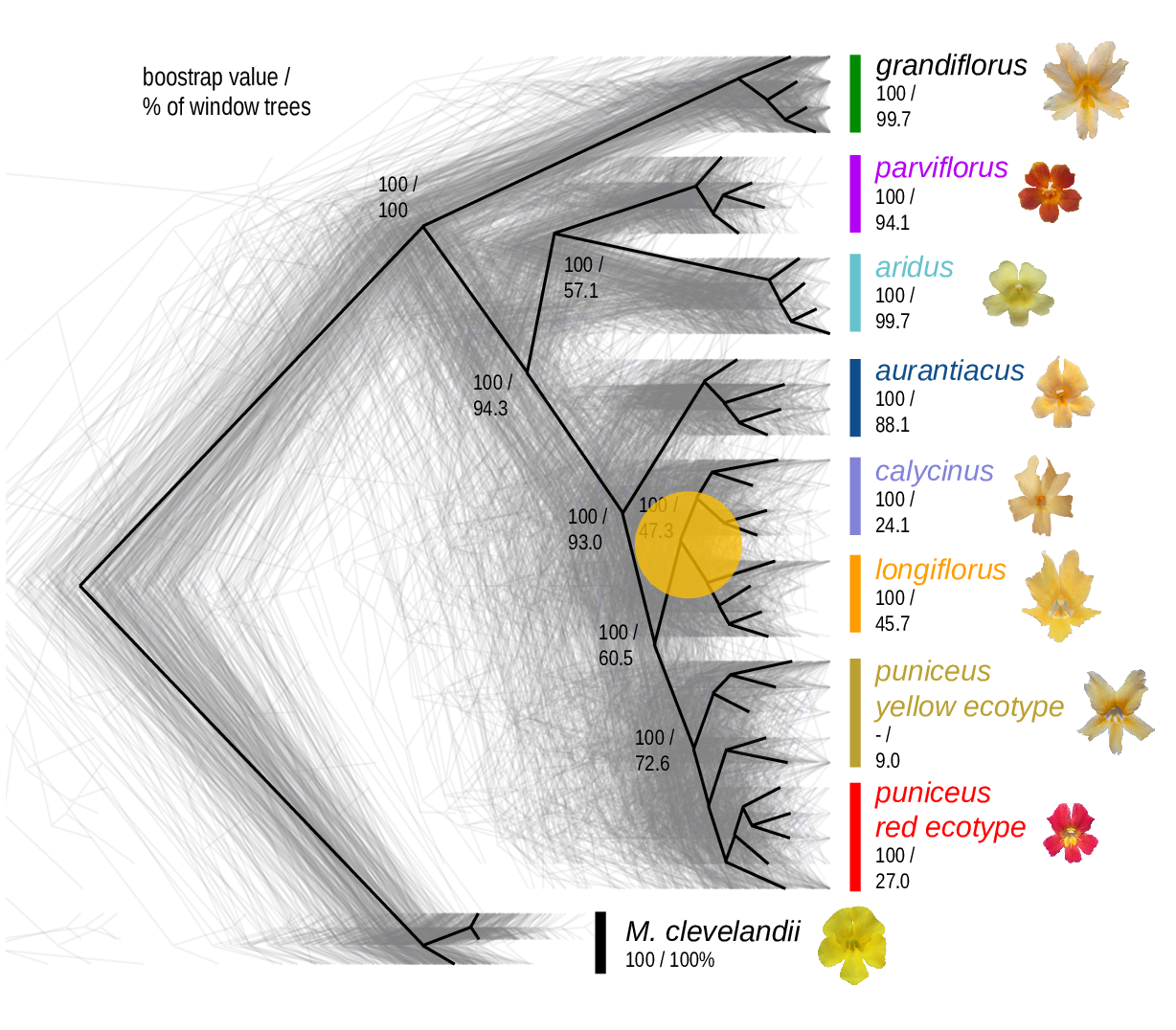
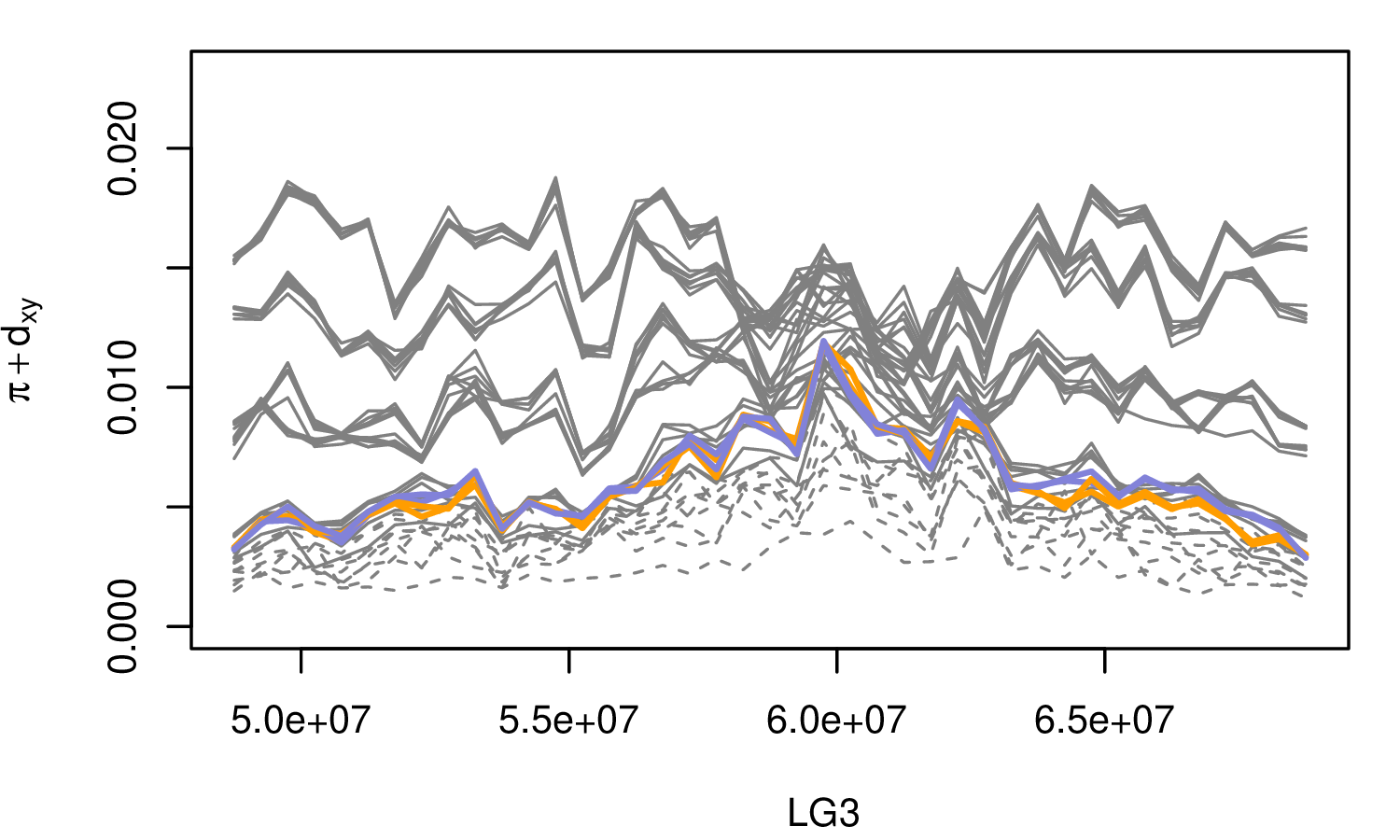
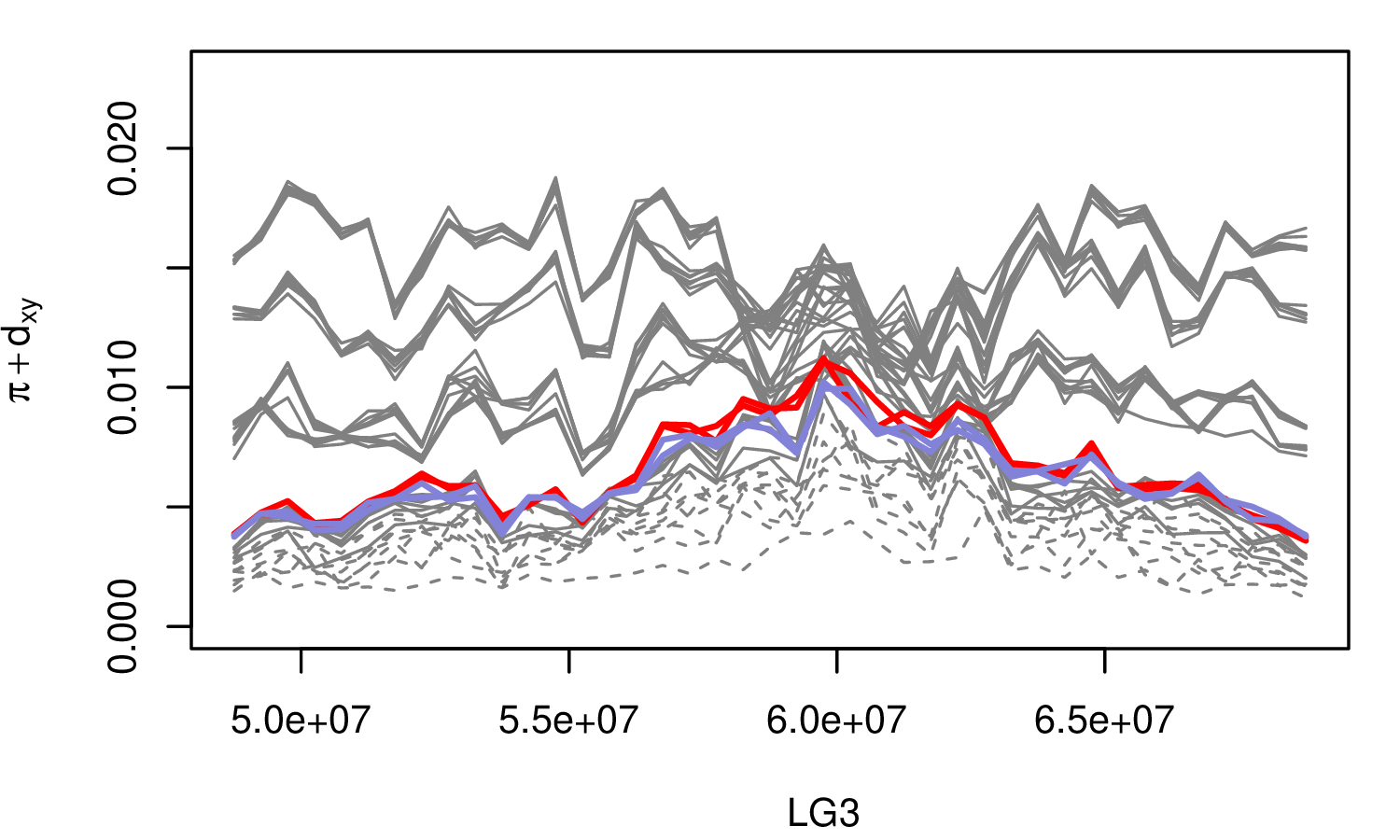
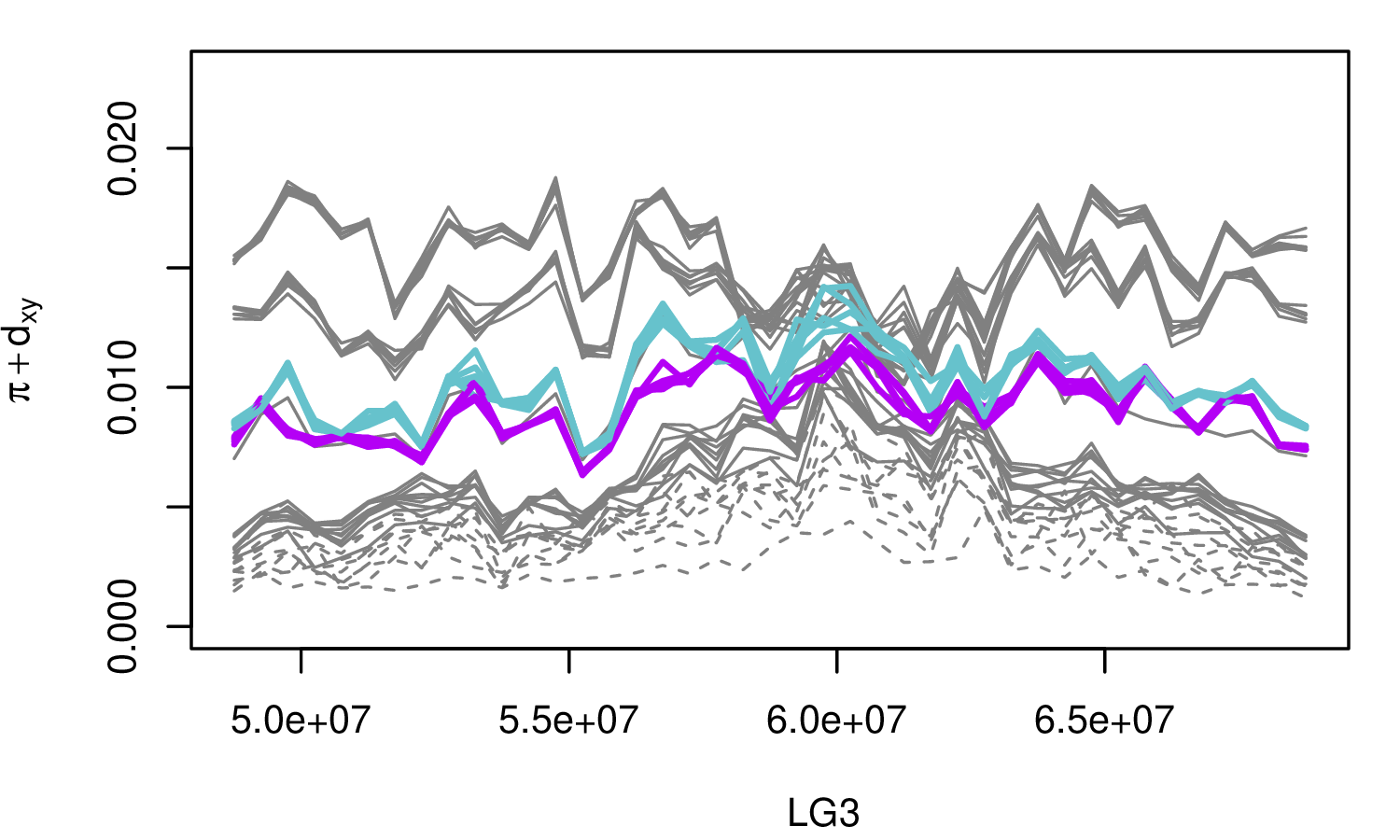
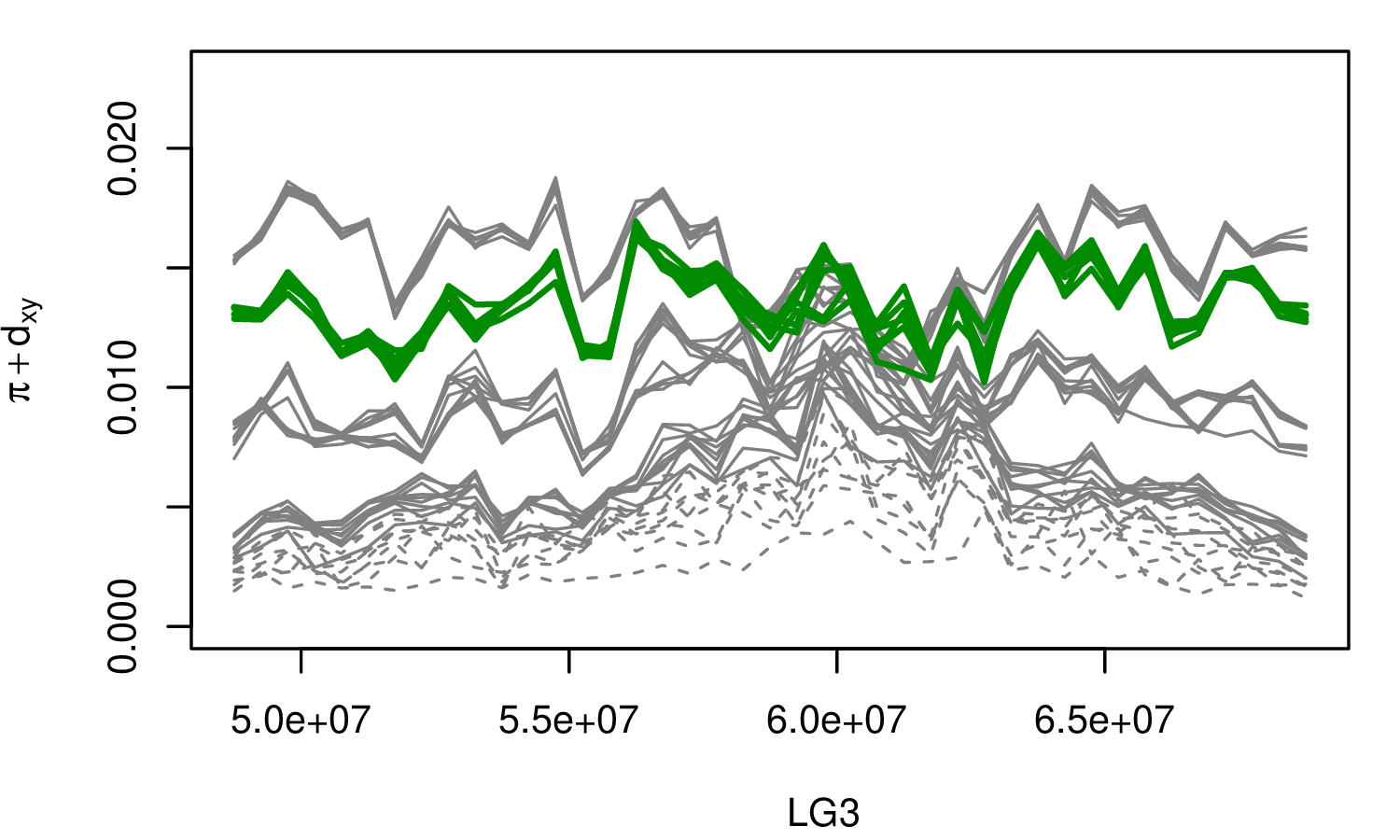
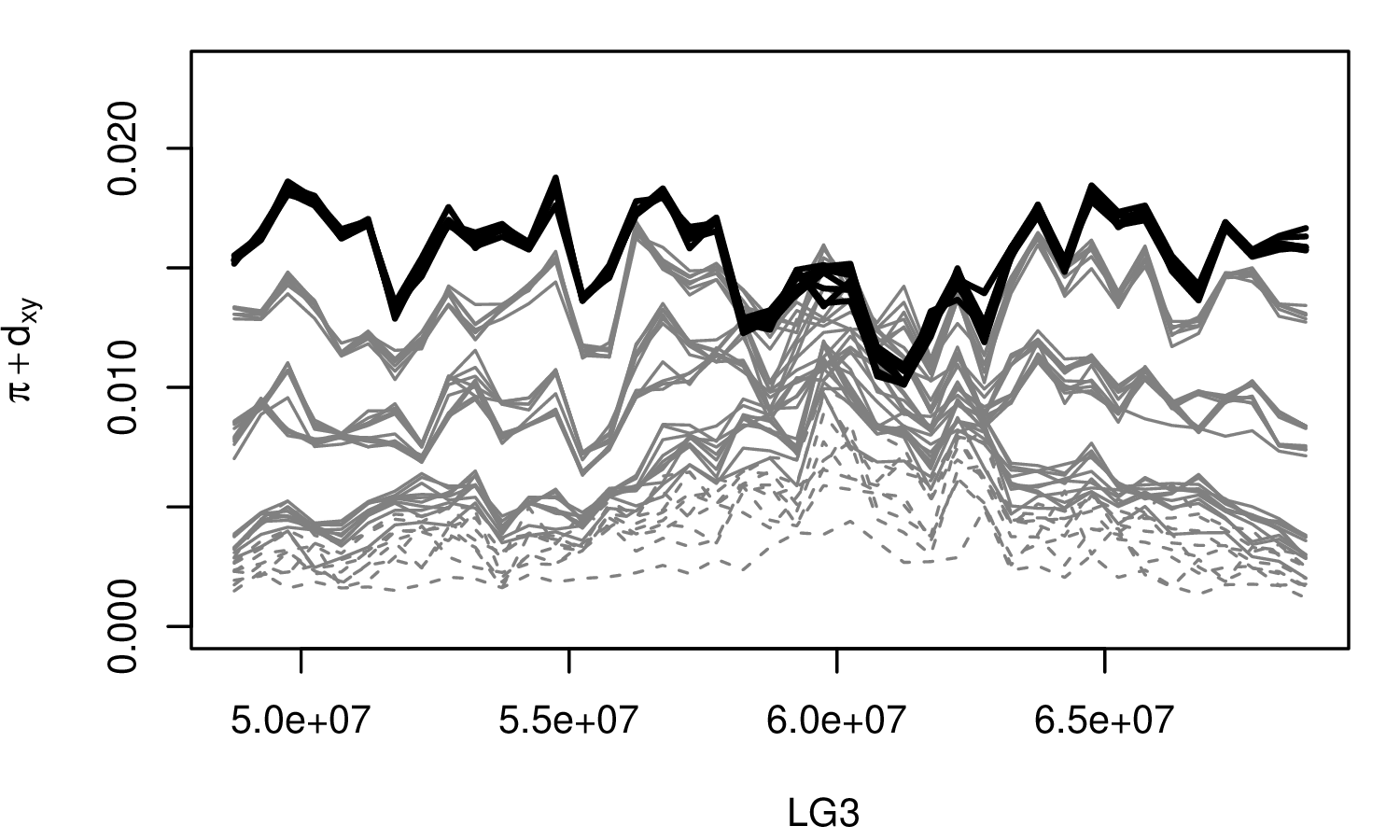
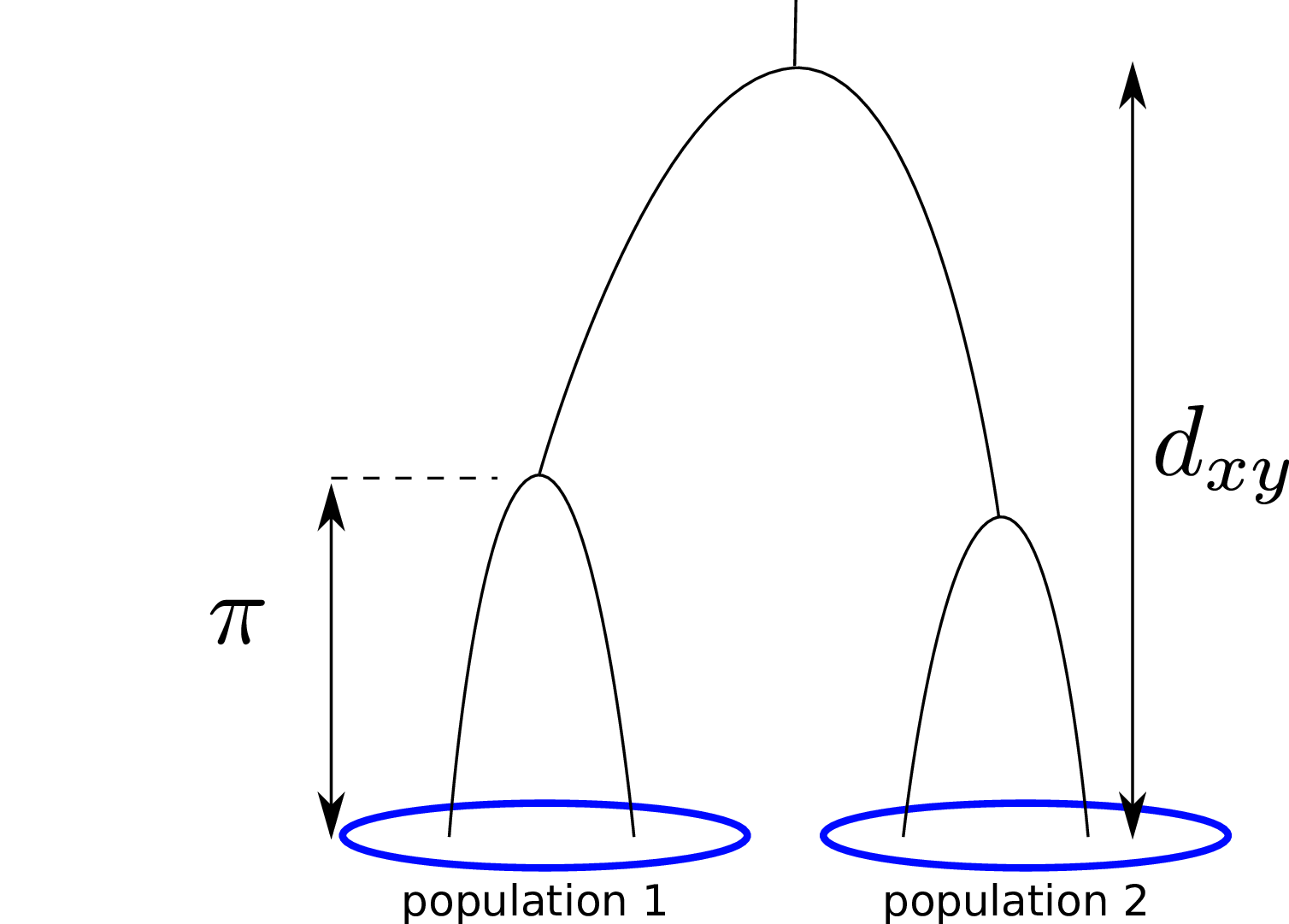
\[ \begin{aligned} \pi &= \text{ (within-pop genetic distance) } \\ d_{xy} &= \text{ (between-pop genetic distance) } \end{aligned} \]
Some questions
How much genetic variation typically underlies traits?
How important is natural selection in determining genetic diversity?
How will populations respond to changes in the future?
To test theories and fit models, we need simulations with realistic
- population sizes,
- genomes,
- selective pressures,
- histories, and
- geography.
The tree sequence
History is a sequence of trees
For a set of sampled chromosomes, at each position along the genome there is a genealogical tree that says how they are related.
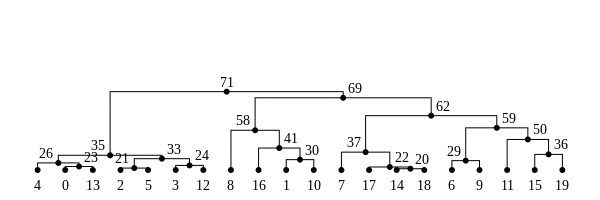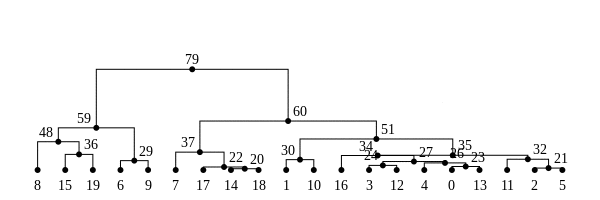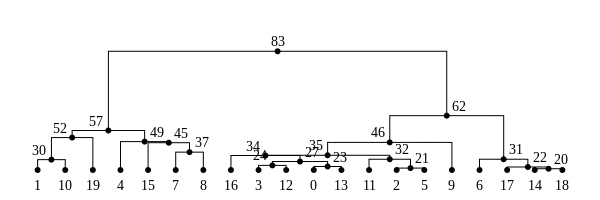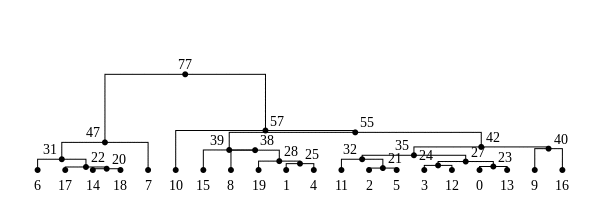
The tree sequence is a way to describe this, er, sequence of trees.
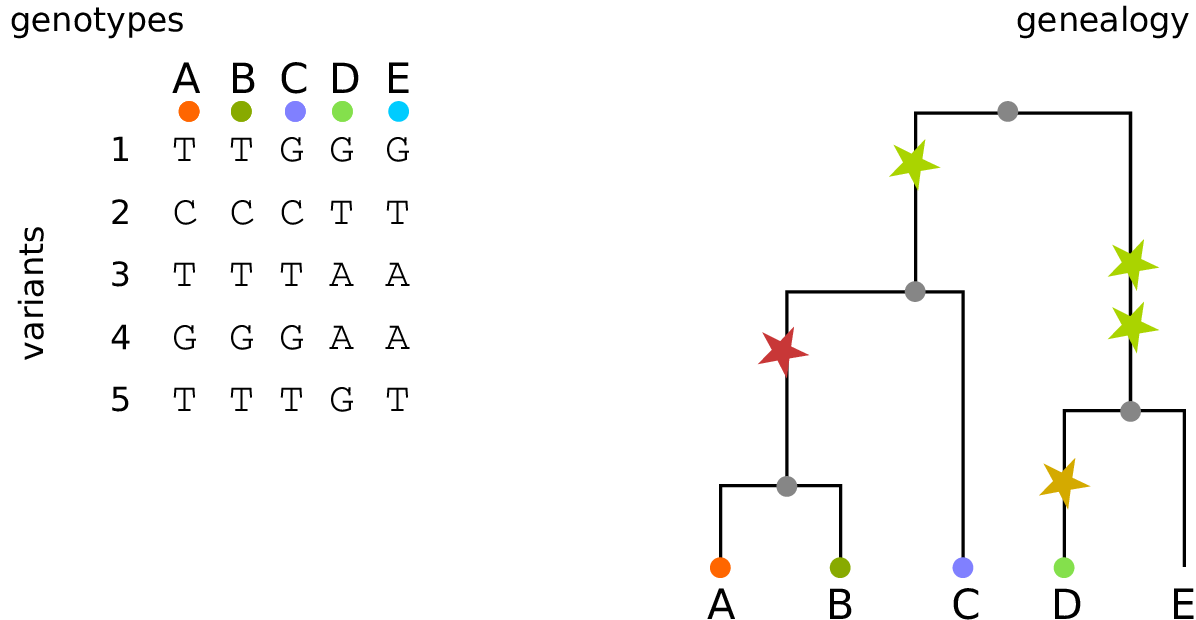
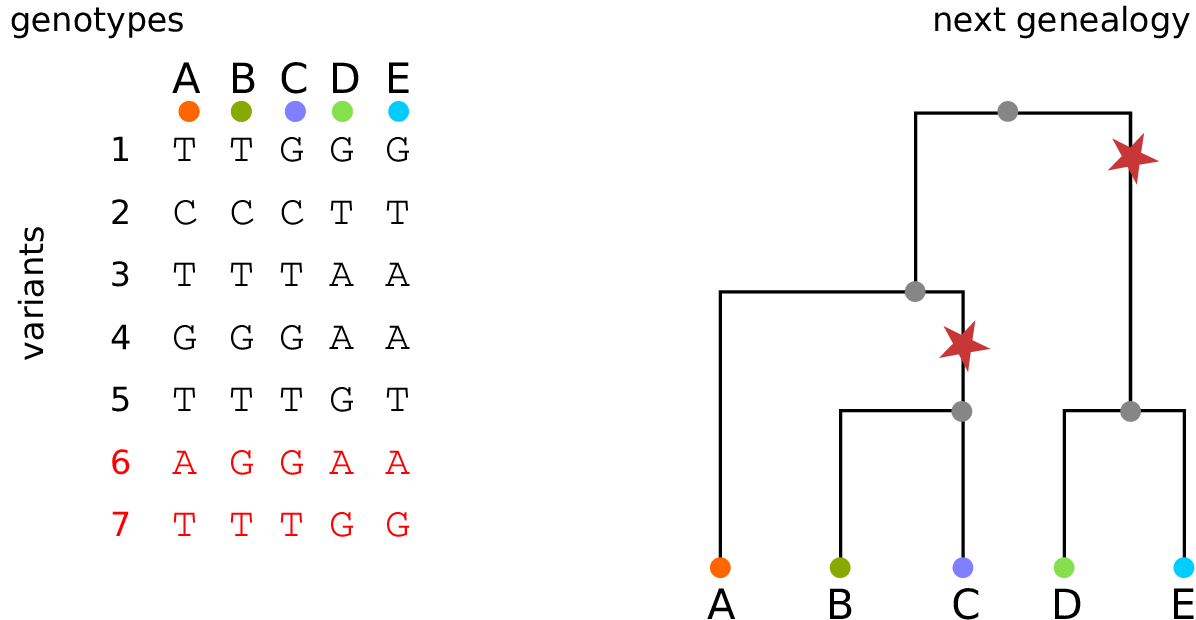
Kelleher, Etheridge, and McVean introduced the tree sequence data structure for a fast coalescent simulator, msprime.
stores sequence and genealogical data very efficiently
tree-based sequence storage closely related to haplotype-matching compression
python/C
tskittools

jerome kelleher
File sizes
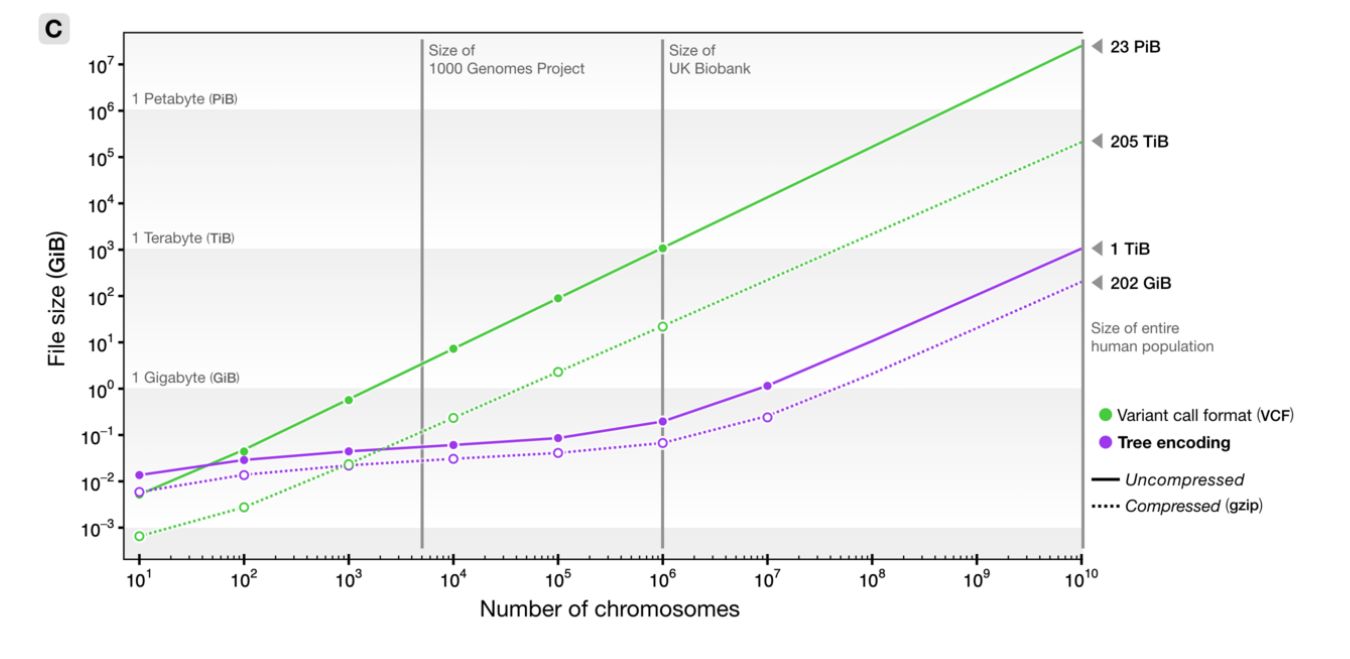
from Kelleher et al 2018, Inferring whole-genome histories in large population datasets, Nature Genetics
Computation run time
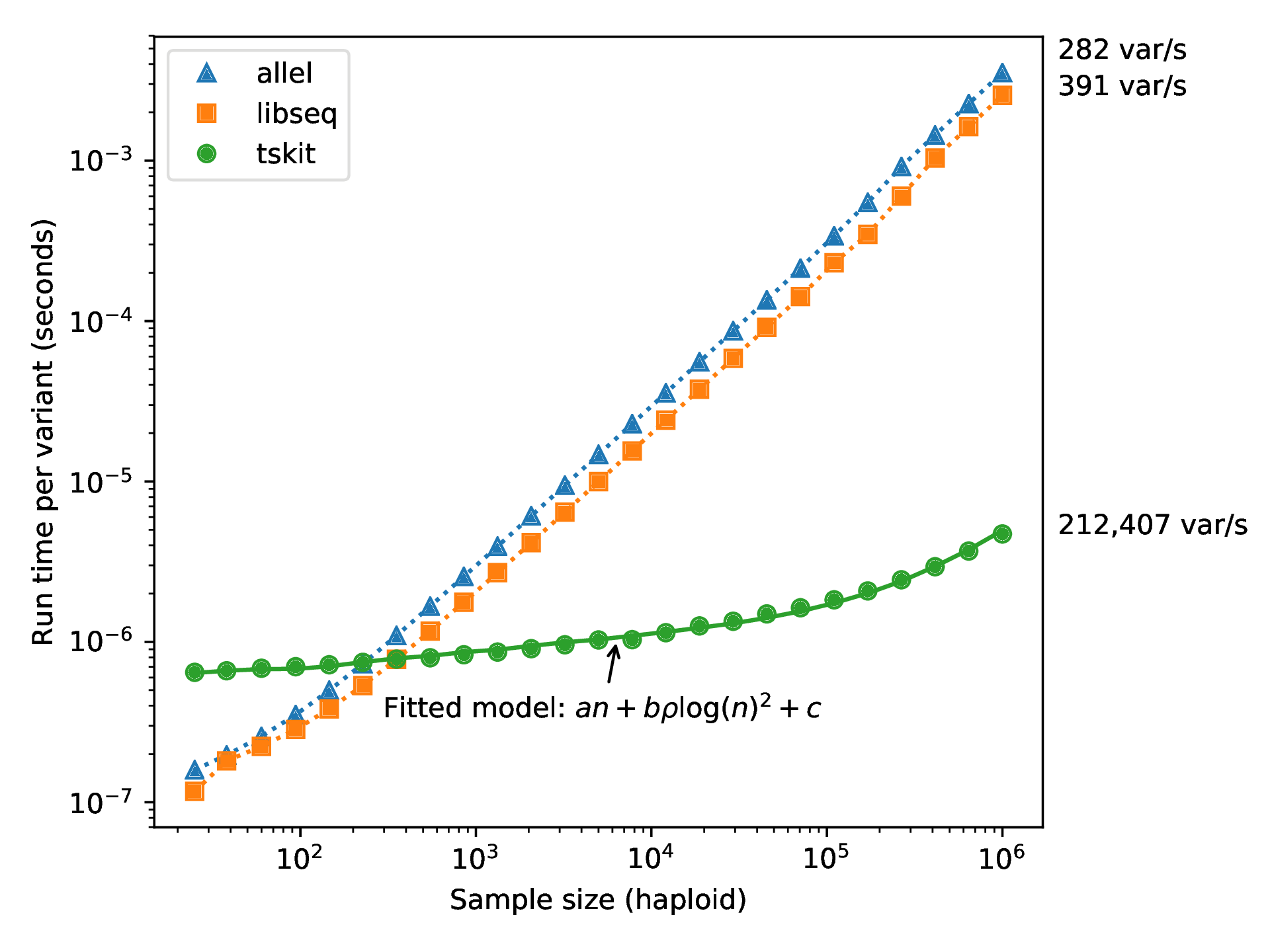
What do genotypes tell us about the genealogies?
Summaries of genotypes and genealogies
Genotypes:
- For each site,
- look at who has which alleles,
- and add a summary of these values to our running total.
Example: genetic distance counts how many mutations differ between two sequences.
Summaries of genotypes and genealogies
Genotypes:
- For each site,
- look at who has inherited which alleles,
- and add a summary of these values to the running total.
Example: sequence divergence counts how many mutations differ between two sequences.
Trees:
- For each branch,
- look at who would inherit mutations on that branch,
- and add the expected contribution to the running total.
Example: the mean time to most recent common ancestor between two sequences.
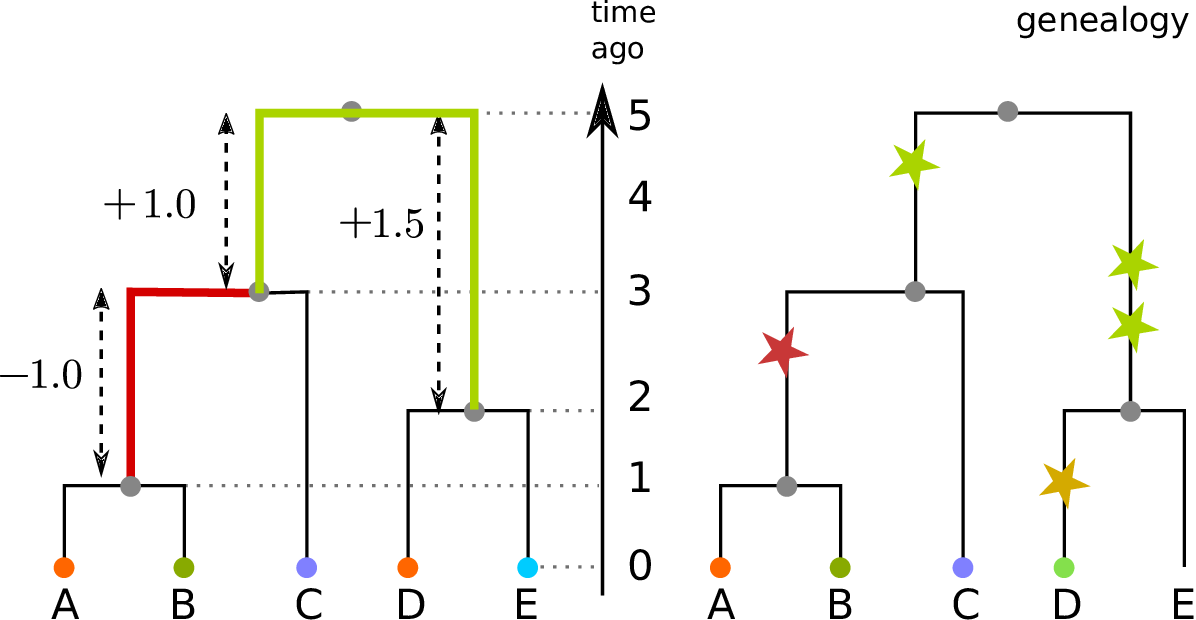
Duality
Any set of sample weights \(w\) and summary function \(f\) defines both
- a statistic of genotypes, \(\text{Site}(f,w)\), and
- a statistic of genealogies, \(\text{Branch}(f,w)\).
With genealogies fixed, and averaging only over mutations with rate \(\mu\), \[\begin{equation} \text{Branch}(f, w) = \frac{1}{\mu} \E\left[ \text{Site}(f, w) \right] . \end{equation}\]
Dealing directly with genealogies can remove the layer of noise due to mutation: \[\begin{equation} \frac{1}{\mu^2} \var\left[\text{Site}(f,w)\right] = \var\left[\text{Branch}(f,w)\right] + \frac{1}{n} \E\left[\text{Branch}(f^2,w)\right] \end{equation}\]
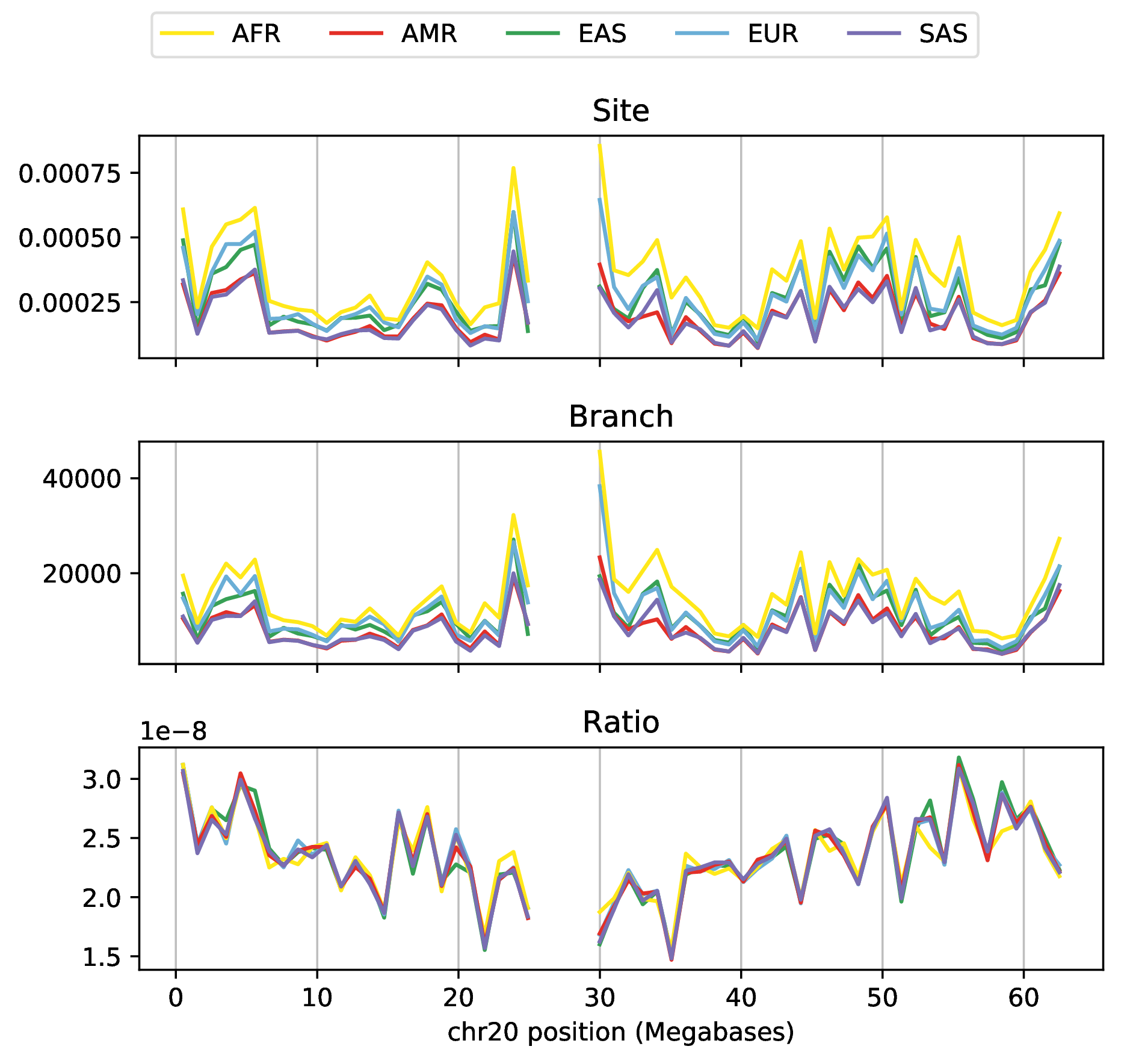
Duality, on 1000 Genomes data? Not quite…
- variation in mutation rate?
- biased gene conversion?
- selection?
- inference artifacts?
Tree sequence from Speidel et al 2019.
Application to demographic inference
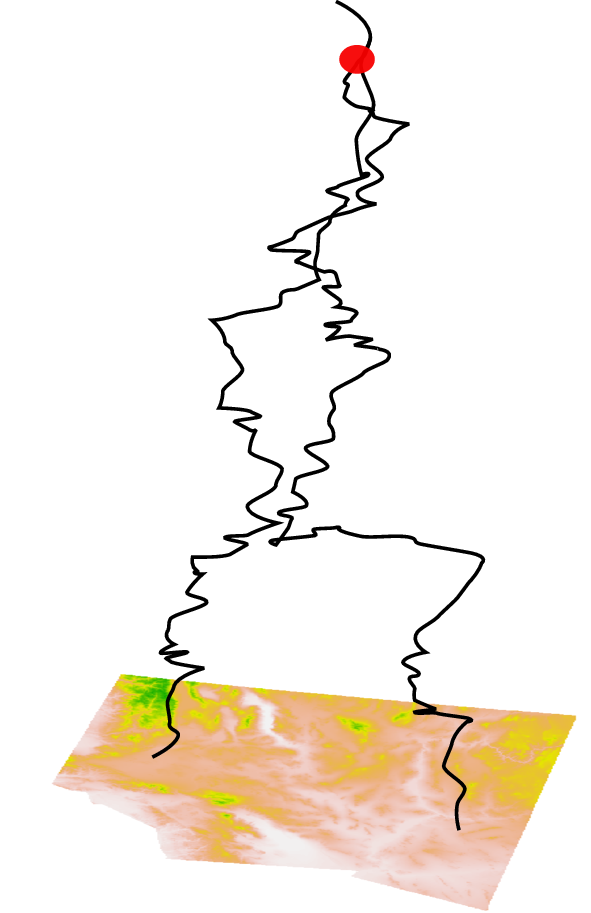
Erik Lundgren: “Isolation By Coalescence”
- fits a discrete random walk model to lineage movement
- genetic distance \(\approx\) mean coalescence time
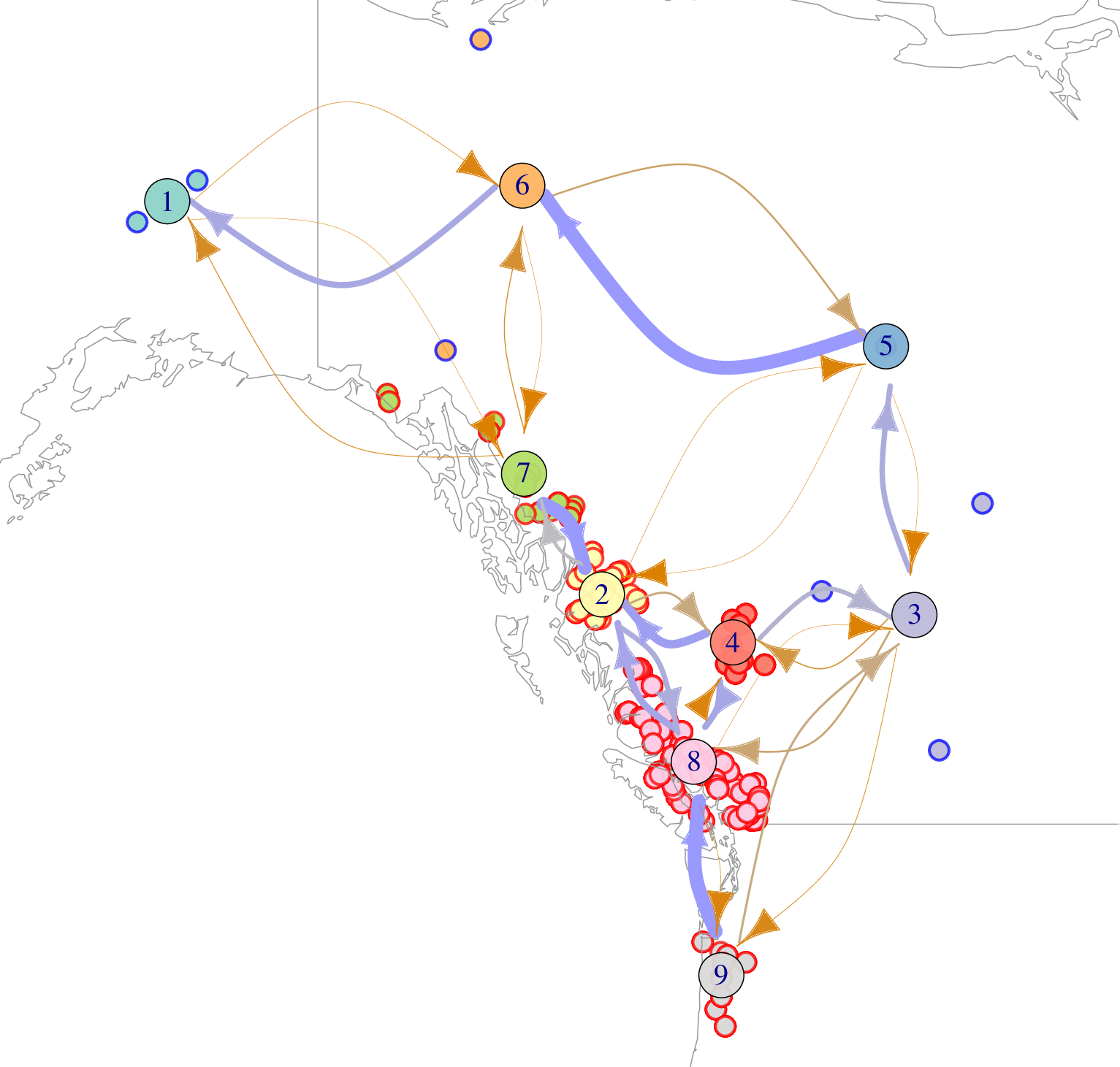
Populus trichocarpa and P. balsamifera data from Moreno Geraldes et al 2014
- glacial refugia
- postglacial expansion
Application to genomic simulations
The main idea
If we record the tree sequence that relates everyone to everyone else,
after the simulation is over we can put neutral mutations down on the trees.
Since neutral mutations don’t affect demography,
this is equivalent to having kept track of them throughout.
From Kelleher, Thornton, Ashander, and Ralph 2018, Efficient pedigree recording for fast population genetics simulation.
and Haller, Galloway, Kelleher, Messer, and Ralph 2018, Tree‐sequence recording in SLiM opens new horizons for forward‐time simulation of whole genomes


This means recording the entire genetic history of everyone in the population, ever.
It is not clear this is a good idea.
But, with a few tricks…
A 100x speedup!
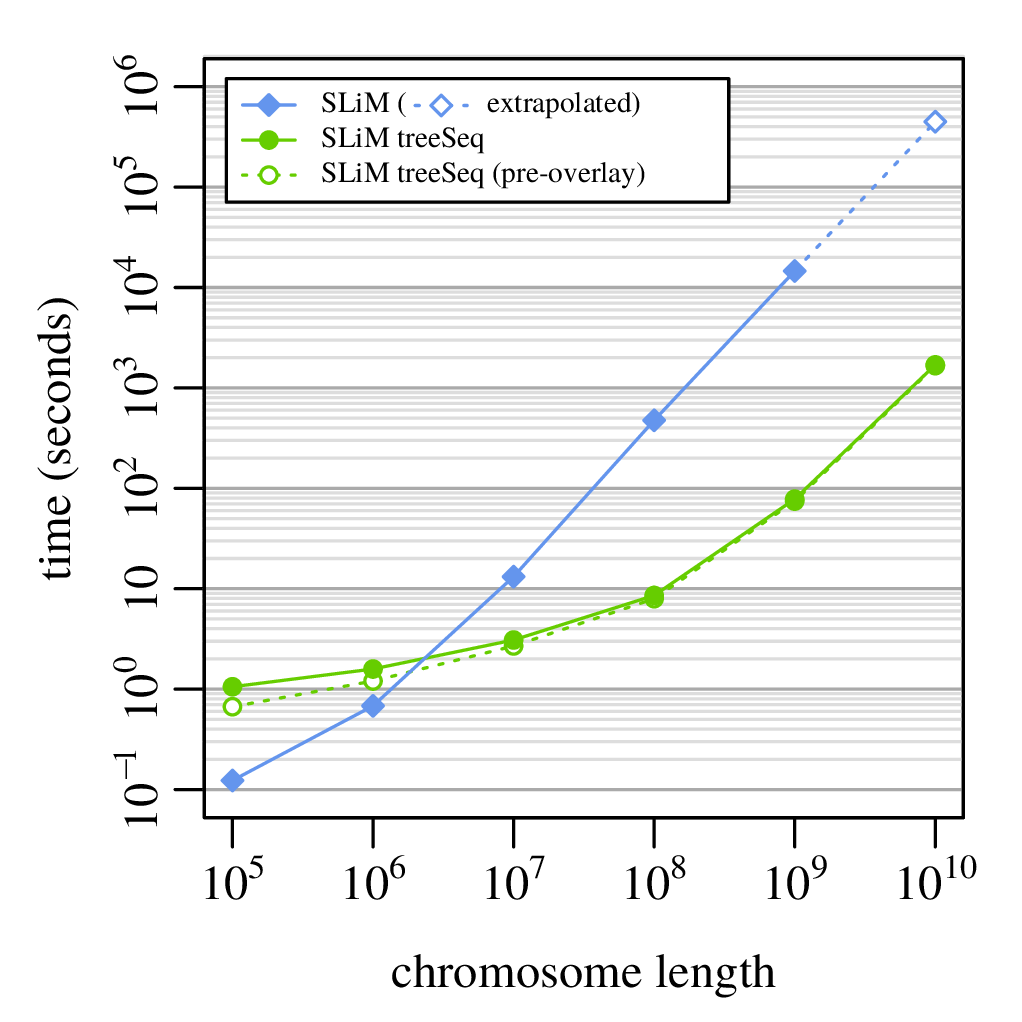
What else can you do with tree sequences?
- record ancient samples
- true ancestry reconstruction
- recapitation: fast, post-hoc initialization with coalescent simulation
For example:
- genome as human chr7 (\(1.54 \times 10^8\)bp)
- \(\approx\) 10,000 diploids
- 500,000 overlapping generations
- continuous, square habitat
- selected mutations at rate \(10^{-10}\)
- neutral mutations added afterwards
Runtime: 8 hours
Back to Mimulus
The data


Simulations
- \(N=10,000\) diploids
- burn-in for \(10N\) generations
population split, with either:
- neutral
- background selection
- selection against introgressed alleles

Murillo Rodrigues
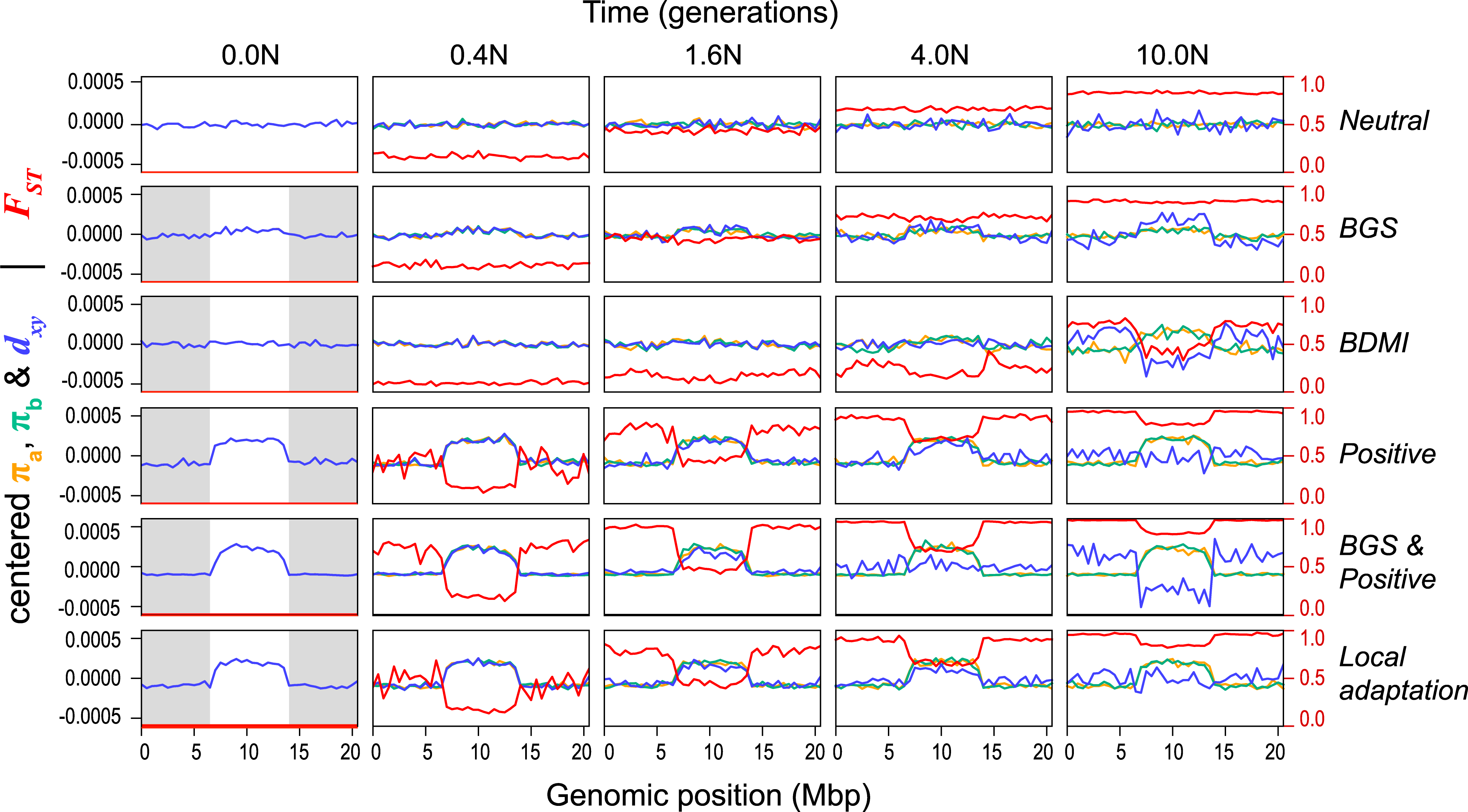
From Widespread selection and gene flow shape the genomic landscape during a radiation of monkeyflowers, Sean Stankowski, Madeline A. Chase, Allison M. Fuiten, Murillo F. Rodrigues, Peter L. Ralph, and Matthew A. Streisfeld; PLoS Bio 2019.
Wrap-up
We need better understanding and prediction of how
- genotype maps to phenotype,
- natural selection acts on phenotypes,
- and that affects genetic variation.
To test theories and fit models, we need simulations with realistic
- population sizes,
- genomes,
- selective pressures,
- histories, and
- geography.
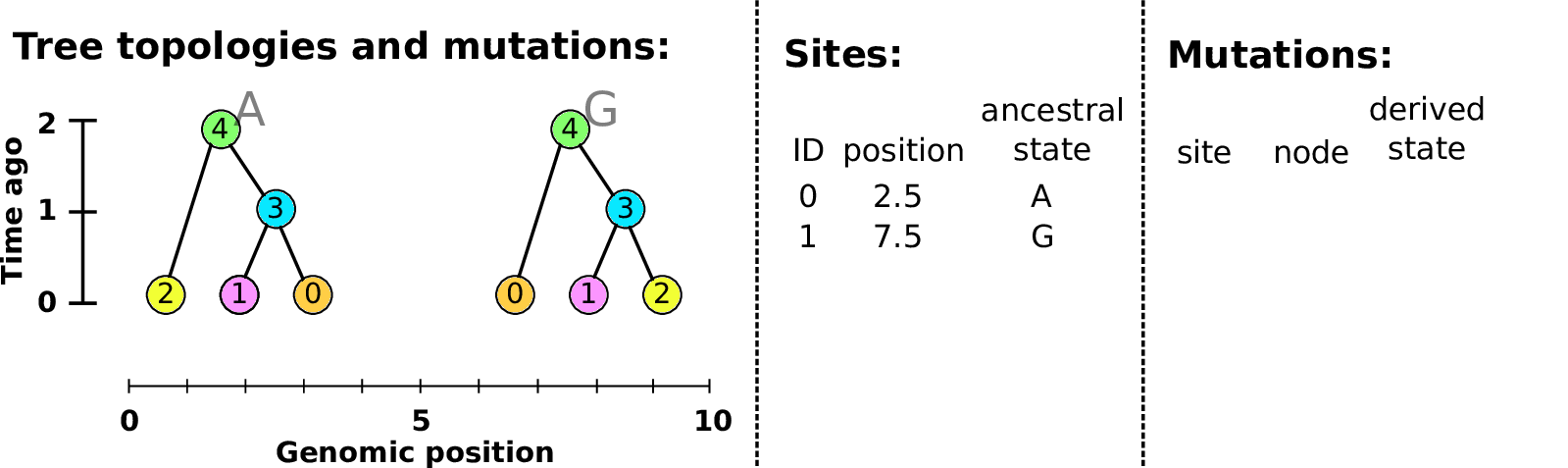
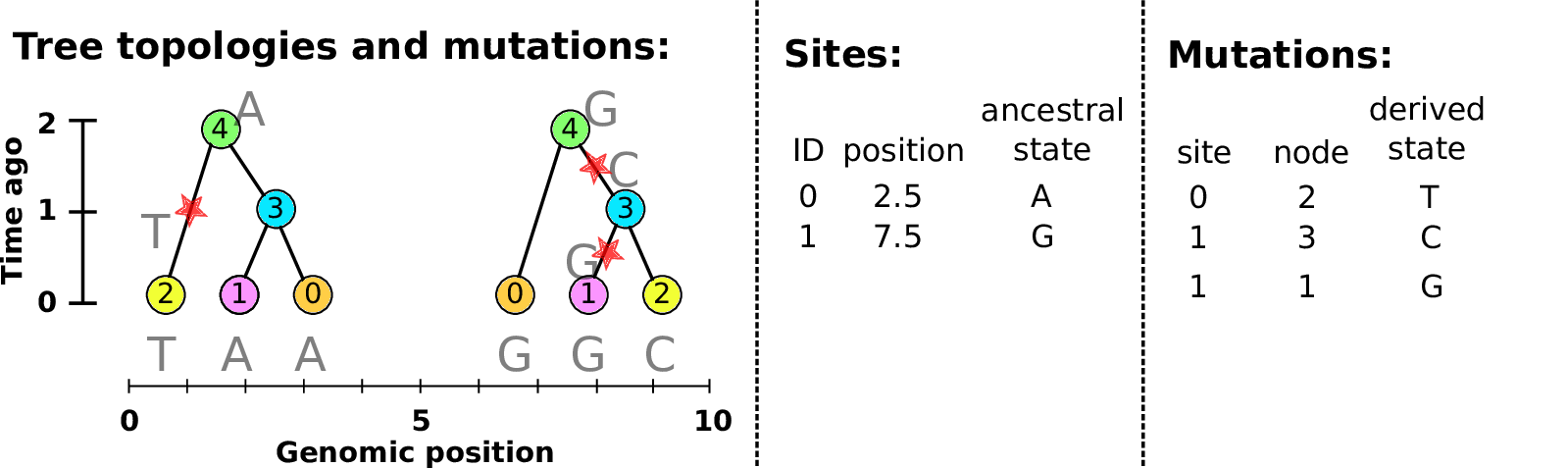
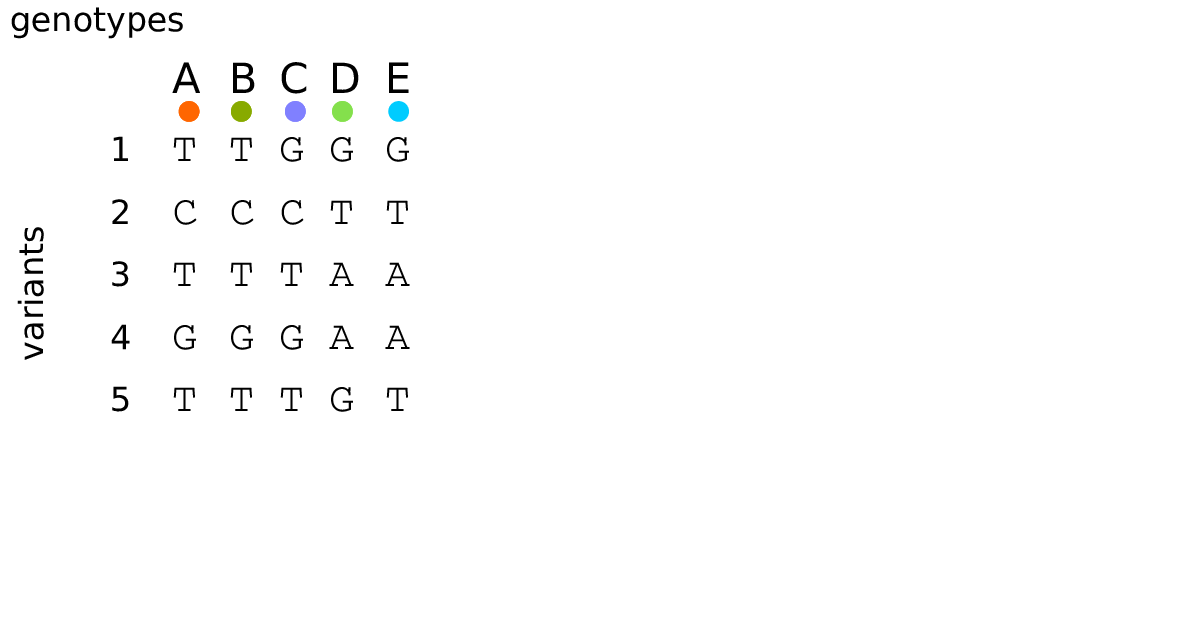
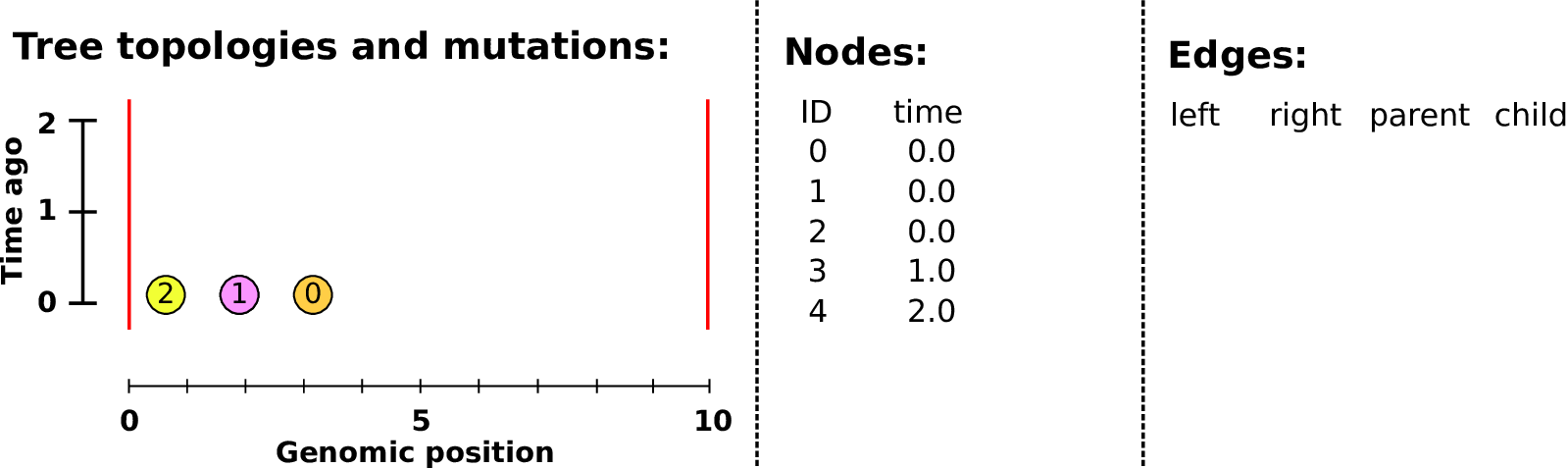
An example tree sequence
Example: three samples; two trees; two variant sites
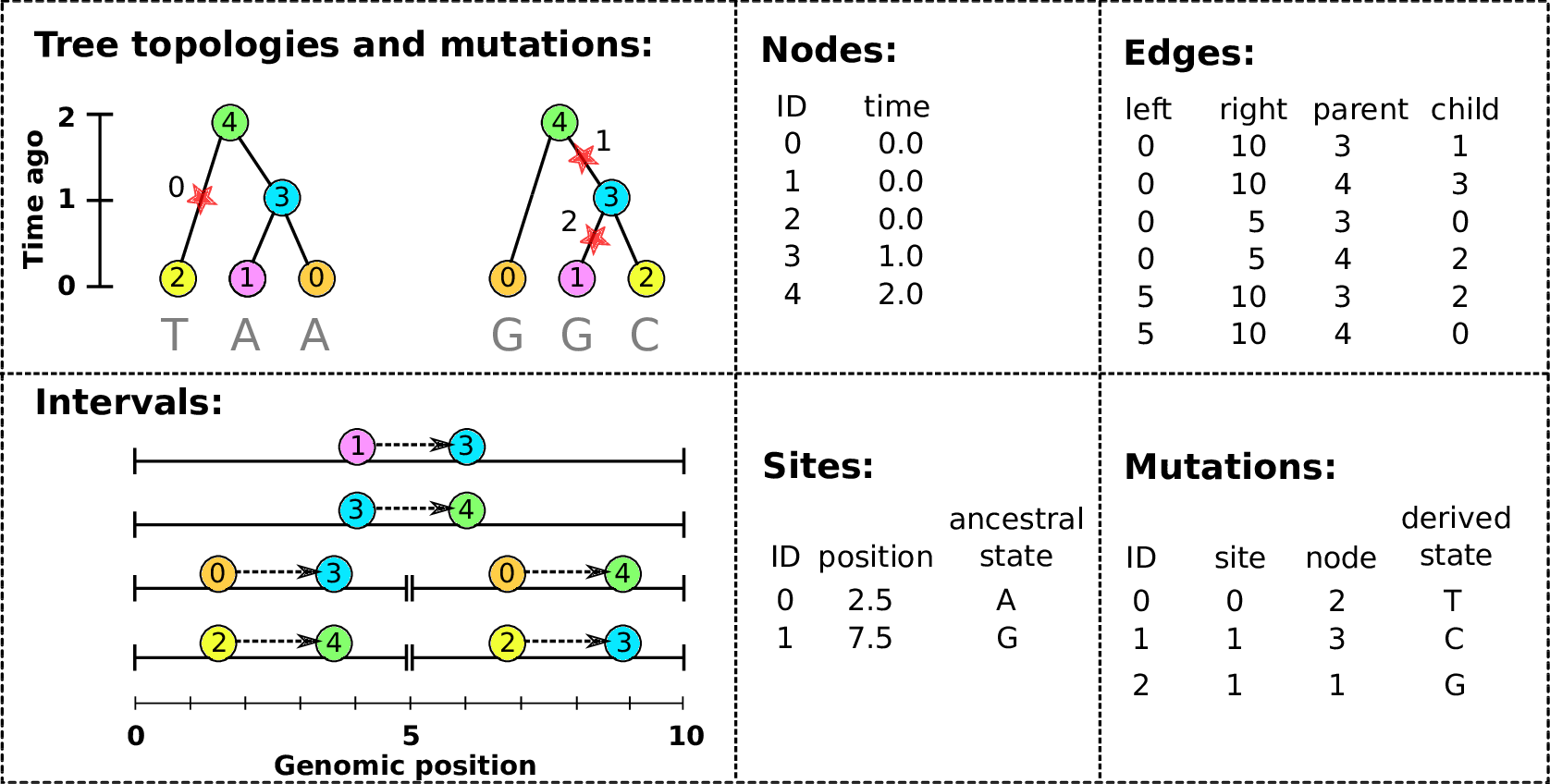
Nodes and edges
- Edges
Who inherits from who; only necessary for coalescent events.
Records: interval (left, right); parent node; child node.
- Nodes
The ancestors those happen in.
Records: time ago (of birth); ID (implicit).
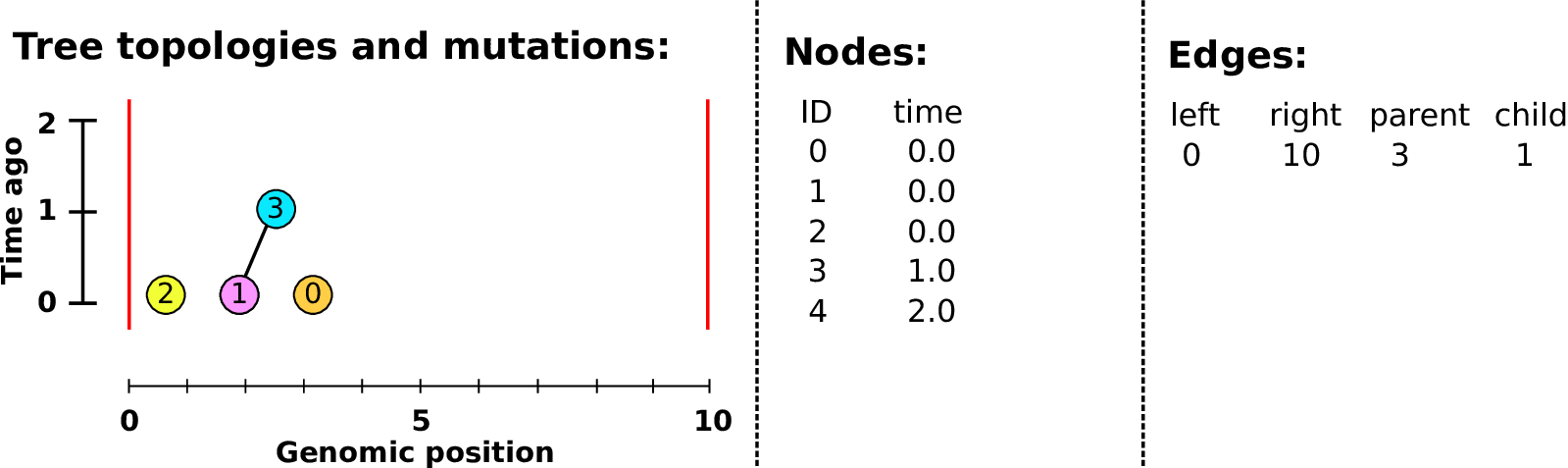
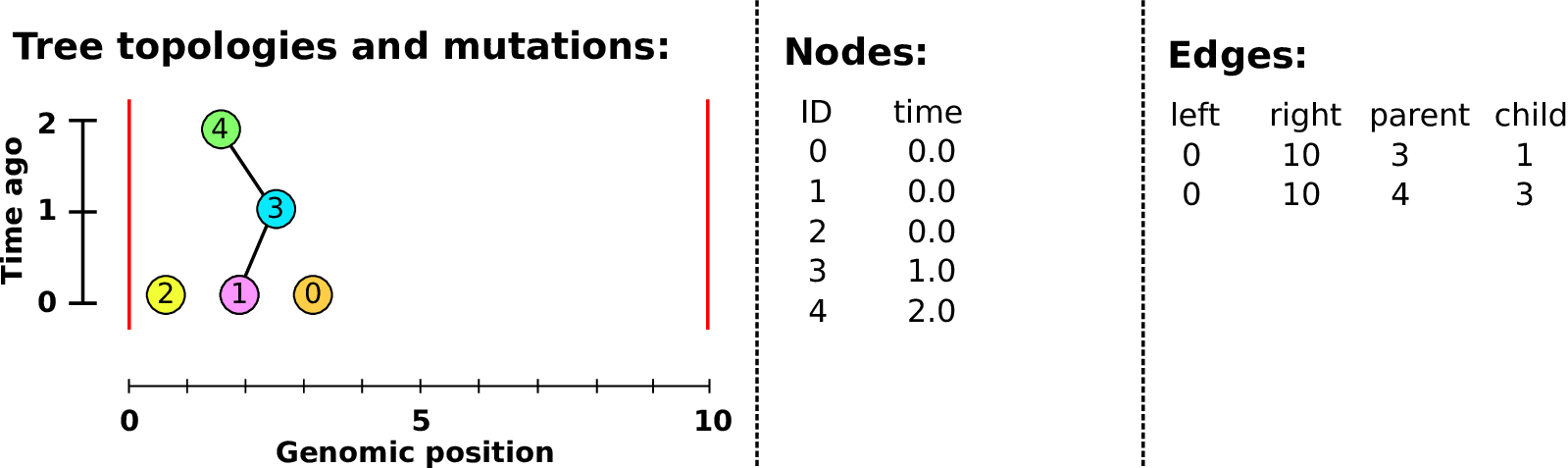
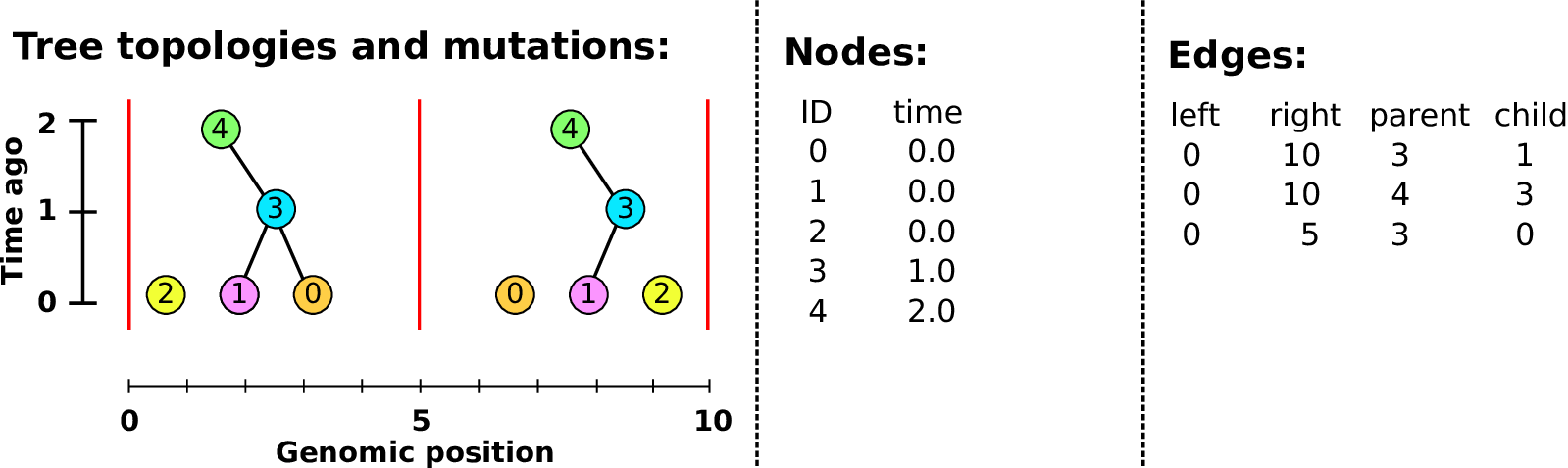
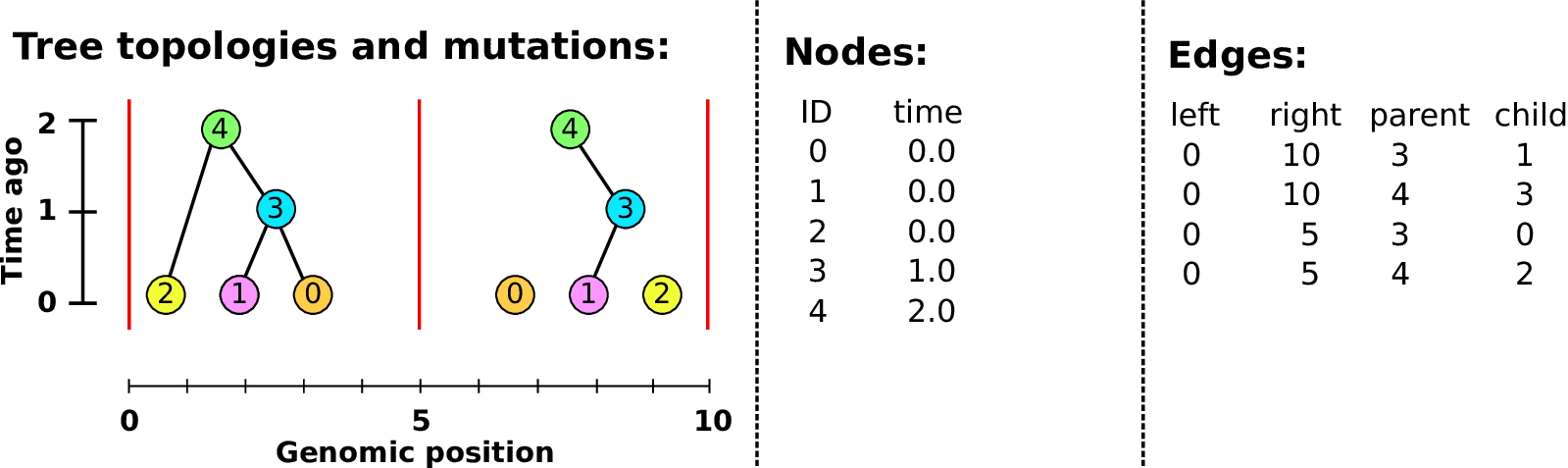
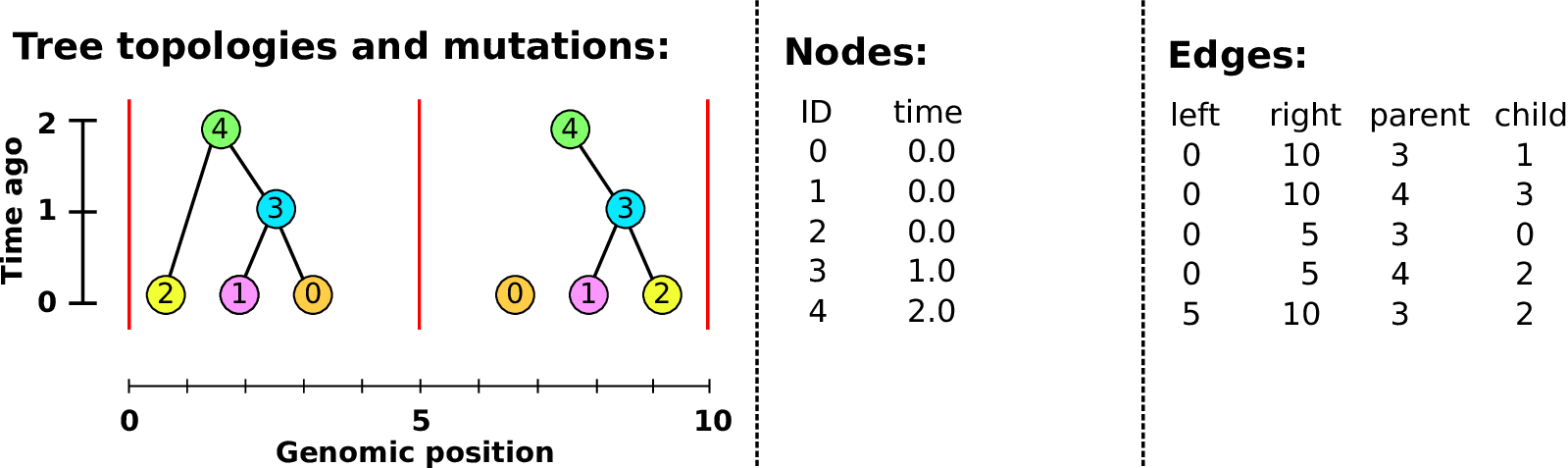
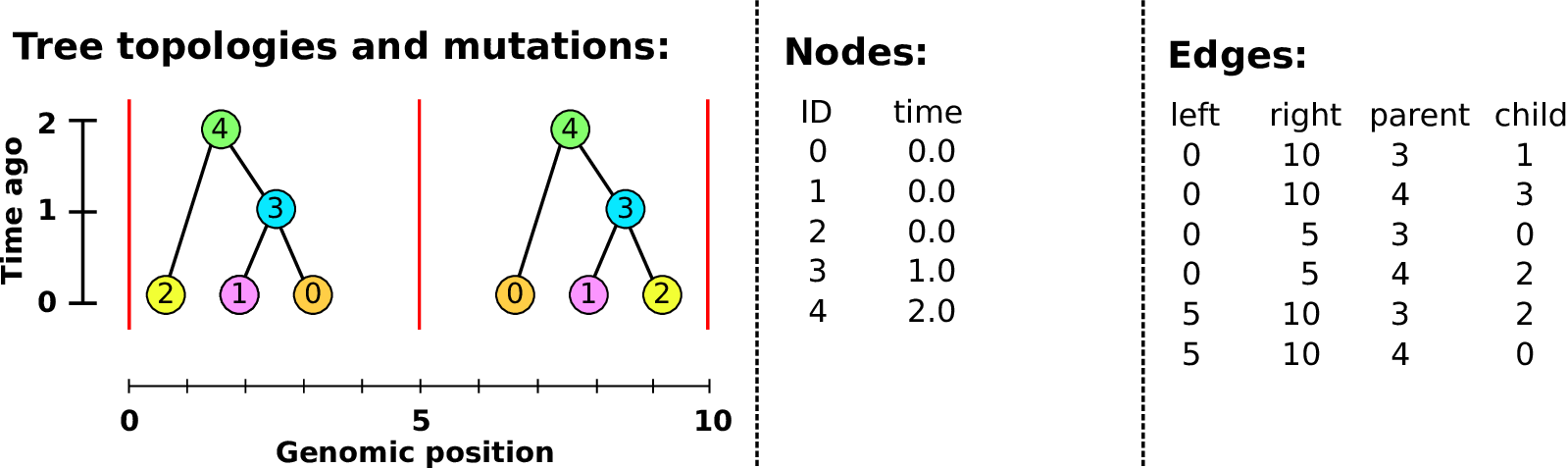
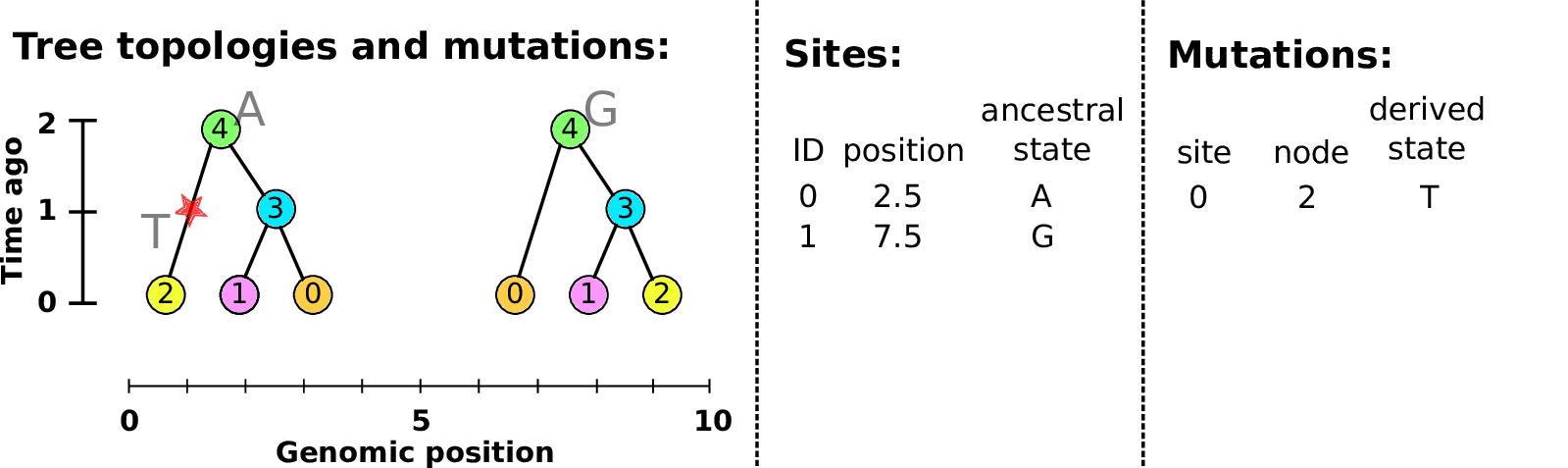
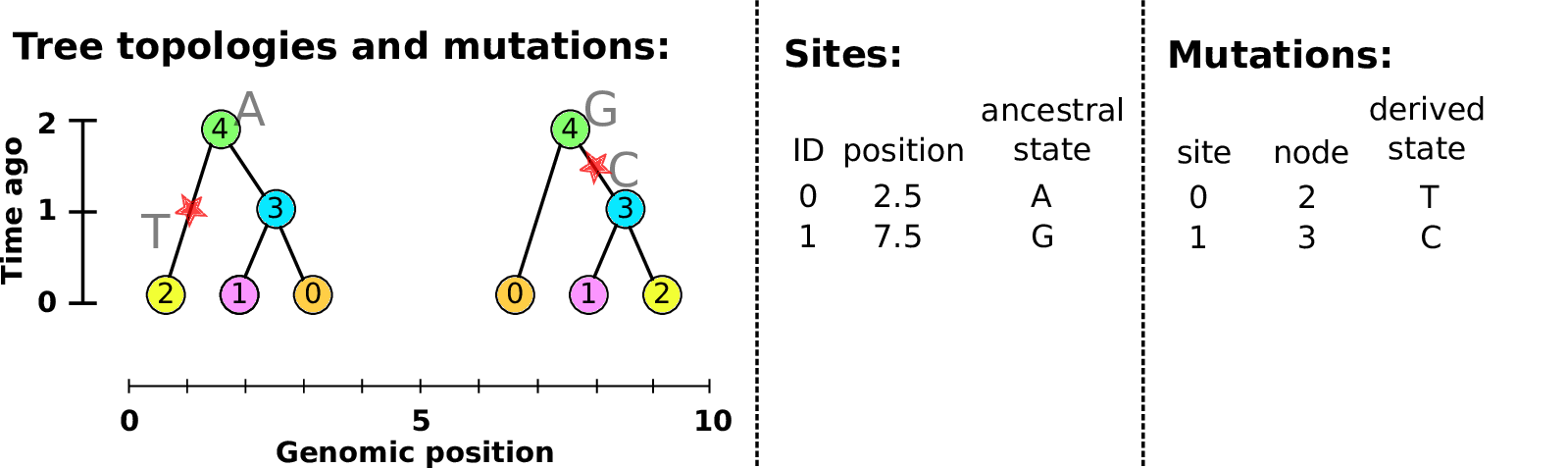
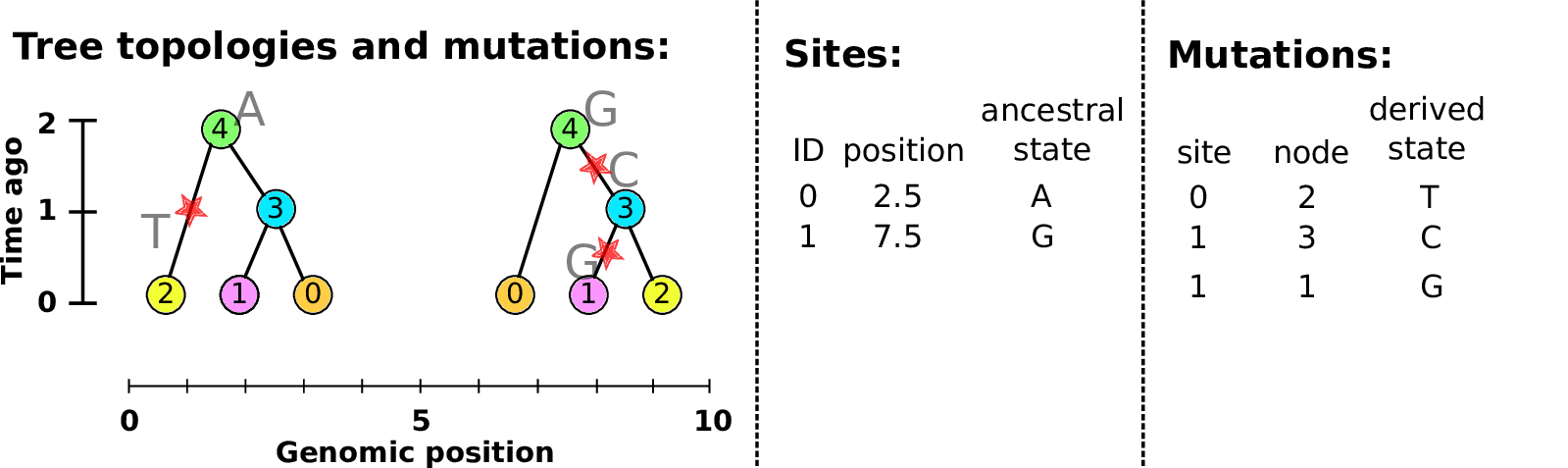
Sites and mutations
- Mutations
When state changes along the tree.
Records: site it occured at; node it occurred in; derived state.
- Sites
Where mutations fall on the genome.
Records: genomic position; ancestral (root) state; ID (implicit).